Modeling X chromosome inactivation using t5iLA naive human pluripotent stem cells
- PMID: 39294757
- PMCID: PMC11411763
- DOI: 10.1186/s12915-024-01994-y
Modeling X chromosome inactivation using t5iLA naive human pluripotent stem cells
Abstract
Background: X chromosome inactivation (XCI) is a critical epigenetic event for dosage compensation of X-linked genes in female mammals, ensuring developmental stability. A robust in vitro model is required for mimicking XCI during the early stages of embryonic development. This methodology article introduces an advanced framework for the in-depth study of XCI using human pluripotent stem cells (hPSCs). By focusing on the transition between naive and primed pluripotent states, we highlight the role of long non-coding RNA X-inactive specific transcript (XIST) and epigenetic alterations in mediating XCI.
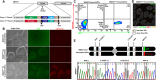
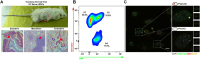
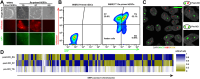
Results: Our methodology enables the distinction between naive and primed hESCs based on XIST expression and the activity of X-linked reporters, facilitating the investigation of XCI initiation and maintenance. Through detailed experimental procedures, we demonstrate the utility of our hESC lines in modeling the process of human XCI, including the establishment of conditions for random XCI induction and the analysis of X chromosome reactivation.
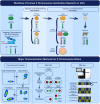
Methods: The study outlines a comprehensive approach for characterizing the X chromosome status in hPSCs, employing dual fluorescent reporter hESC lines. These reporter lines enable real-time tracking of XCI dynamics through differentiation processes. We detailed protocols for the induction of X chromosome reactivation and inactivation, as well as the X status characterization methods including cultivation of hESCs, flow cytometric analysis, RNA fluorescence in situ hybridization (FISH), and transcriptome sequencing, providing a step-by-step guide for researchers to investigate XCI mechanisms in vitro.
Conclusions: This article provides a detailed, reproducible methodology for studying XCI mechanisms in vitro, employing hPSCs as a model system. It presents a significant advance in our ability to investigate XCI, offering potential applications in developmental biology, disease modeling, and regenerative medicine. By facilitating the study of XCI dynamics, this methodological framework paves the way for deeper understanding and manipulation of this fundamental biological process.
Keywords: Early embryogenesis; Human embryonic stem cells; X chromosome inactivation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Female human pluripotent stem cells rapidly lose X chromosome inactivation marks and progress to a skewed methylation pattern during culture.Mol Hum Reprod. 2016 Apr;22(4):285-98. doi: 10.1093/molehr/gaw004. Epub 2016 Jan 19. Mol Hum Reprod. 2016. PMID: 26786180
-
Global Characterization of X Chromosome Inactivation in Human Pluripotent Stem Cells.Cell Rep. 2019 Apr 2;27(1):20-29.e3. doi: 10.1016/j.celrep.2019.03.019. Cell Rep. 2019. PMID: 30943402
-
Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation.Cell Stem Cell. 2017 Jan 5;20(1):87-101. doi: 10.1016/j.stem.2016.10.006. Epub 2016 Dec 15. Cell Stem Cell. 2017. PMID: 27989770 Free PMC article.
-
X chromosome inactivation in human pluripotent stem cells as a model for human development: back to the drawing board?Hum Reprod Update. 2017 Sep 1;23(5):520-532. doi: 10.1093/humupd/dmx015. Hum Reprod Update. 2017. PMID: 28582519 Review.
-
Female-bias in systemic lupus erythematosus: How much is the X chromosome to blame?Biol Sex Differ. 2024 Oct 7;15(1):76. doi: 10.1186/s13293-024-00650-y. Biol Sex Differ. 2024. PMID: 39375734 Free PMC article. Review.
References
-
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, et al. Establishment of Histone H3 Methylation on the Inactive X Chromosome Requires Transient Recruitment of Eed-Enx1 Polycomb Group Complexes. Dev Cell. 2003;4(4):481–95. - PubMed
-
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–5. - PubMed
-
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–76. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

