The genome sequence of the Brown Litter Worm, Bimastos eiseni (Levinsen, 1884)
- PMID: 39296368
- PMCID: PMC11408909
- DOI: 10.12688/wellcomeopenres.21622.1
The genome sequence of the Brown Litter Worm, Bimastos eiseni (Levinsen, 1884)
Abstract
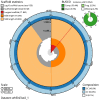
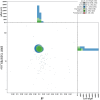
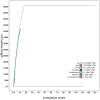
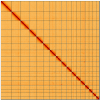
We present a genome assembly from an individual Bimastos eiseni (the Brown Litter Worm; Annelida; None; Haplotaxida; Lumbricidae). The genome sequence is 660.5 megabases in span. Most of the assembly is scaffolded into 17 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 15.34 kilobases in length.
Keywords: Bimastos eiseni; Brown Litter Worm; Haplotaxida; chromosomal; genome sequence.
Copyright: © 2024 Brown KD et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
The genome sequence of the common earthworm, Lumbricus terrestris (Linnaeus, 1758).Wellcome Open Res. 2023 Oct 30;8:500. doi: 10.12688/wellcomeopenres.20178.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 38249959 Free PMC article.
-
The genome sequence of the red compost earthworm, Lumbricus rubellus (Hoffmeister, 1843).Wellcome Open Res. 2023 Aug 18;8:354. doi: 10.12688/wellcomeopenres.19834.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 38618197 Free PMC article.
-
The genome sequence of the mottled worm, Allolobophora icterica (formerly Aporrectodea icterica ) (Savigny, 1826).Wellcome Open Res. 2024 Sep 26;9:556. doi: 10.12688/wellcomeopenres.23066.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39494197 Free PMC article.
-
The genome sequence of a scale worm, Harmothoe impar (Johnston, 1839).Wellcome Open Res. 2023 Jul 20;8:315. doi: 10.12688/wellcomeopenres.19570.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37786780 Free PMC article.
-
The genome sequence of the scale worm, Lepidonotus clava (Montagu, 1808).Wellcome Open Res. 2022 Dec 21;7:307. doi: 10.12688/wellcomeopenres.18660.1. eCollection 2022. Wellcome Open Res. 2022. PMID: 37362008 Free PMC article.
Cited by
-
Annelid Comparative Genomics and the Evolution of Massive Lineage-Specific Genome Rearrangement in Bilaterians.Mol Biol Evol. 2024 Sep 4;41(9):msae172. doi: 10.1093/molbev/msae172. Mol Biol Evol. 2024. PMID: 39141777 Free PMC article.
References
LinkOut - more resources
Full Text Sources

