Variation of tRNA modifications with and without intron dependency
- PMID: 39296543
- PMCID: PMC11408192
- DOI: 10.3389/fgene.2024.1460902
Variation of tRNA modifications with and without intron dependency
Abstract
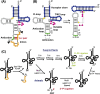
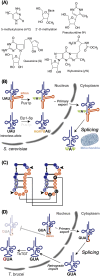
tRNAs have recently gained attention for their novel regulatory roles in translation and for their diverse functions beyond translation. One of the most remarkable aspects of tRNA biogenesis is the incorporation of various chemical modifications, ranging from simple base or ribose methylation to more complex hypermodifications such as formation of queuosine and wybutosine. Some tRNAs are transcribed as intron-containing pre-tRNAs. While the majority of these modifications occur independently of introns, some are catalyzed in an intron-inhibitory manner, and in certain cases, they occur in an intron-dependent manner. This review focuses on pre-tRNA modification, including intron-containing pre-tRNA, in both intron-inhibitory and intron-dependent fashions. Any perturbations in the modification and processing of tRNAs may lead to a range of diseases and disorders, highlighting the importance of understanding these mechanisms in molecular biology and medicine.
Keywords: RNA modification; enzyme; intron; pre-tRNA; processing; tRNA.
Copyright © 2024 Hayashi.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review.Biochimie. 1997 May;79(5):293-302. doi: 10.1016/s0300-9084(97)83517-1. Biochimie. 1997. PMID: 9258438 Review.
-
To be or not to be modified: Miscellaneous aspects influencing nucleotide modifications in tRNAs.IUBMB Life. 2019 Aug;71(8):1126-1140. doi: 10.1002/iub.2041. Epub 2019 Apr 1. IUBMB Life. 2019. PMID: 30932315 Free PMC article. Review.
-
Intron-Dependent or Independent Pseudouridylation of Precursor tRNA Containing Atypical Introns in Cyanidioschyzon merolae.Int J Mol Sci. 2022 Oct 11;23(20):12058. doi: 10.3390/ijms232012058. Int J Mol Sci. 2022. PMID: 36292915 Free PMC article.
-
Transfer RNA processing - from a structural and disease perspective.Biol Chem. 2022 Jun 21;403(8-9):749-763. doi: 10.1515/hsz-2021-0406. Print 2022 Jul 26. Biol Chem. 2022. PMID: 35728022 Review.
-
Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo.Mol Cell Biol. 1986 Jul;6(7):2663-73. doi: 10.1128/mcb.6.7.2663-2673.1986. Mol Cell Biol. 1986. PMID: 3537724 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

