Golgi clustering by the deficiency of COPI-SNARE in Drosophila photoreceptors
- PMID: 39296936
- PMCID: PMC11408282
- DOI: 10.3389/fcell.2024.1442198
Golgi clustering by the deficiency of COPI-SNARE in Drosophila photoreceptors
Abstract
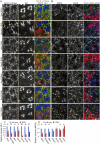
A comprehensive study of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) in the fly genome by RNAi in Drosophila photoreceptors indicated that knockdown of any of the COPI-SNAREs, Syx18, Sec20, and Use1, resulted in the same characteristic phenotypes: Golgi stacks gathering on their trans-side, laterally expanded Golgi cisternae, and a reduced number of discrete Golgi stacks. These Golgi stacks are reminiscent of mammalian Golgi ribbons and Brefeldin A (BFA)-bodies in Drosophila S2 cells. As previously reported, BFA suppresses trans-Golgi network (TGN) fission and Golgi stack separation to form a BFA-body, which is a cluster of Golgi stacks cored by recycling endosomes. We found that the impairing each of COPI-SNAREs results in clustered Golgi stacks similar to BFA-bodies, indicating that COPI-SNAREs have a role to separate clustered Golgi stacks. These results further support the idea that the movement of Golgi stacks and the balance of fusion and fission of the TGN determine the level of clustering and ribbon formation of Golgi stacks within cells.
Keywords: BFA-body; Drosophila; Golgi stacks; photoreceptors; recycling endosomes.
Copyright © 2024 Tago, Yamada, Goto, Toyooka, Ochi, Satoh and Satoh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

