Genome-wide analysis of the ERF Family in Stephania japonica provides insights into the regulatory role in Cepharanthine biosynthesis
- PMID: 39297007
- PMCID: PMC11408324
- DOI: 10.3389/fpls.2024.1433015
Genome-wide analysis of the ERF Family in Stephania japonica provides insights into the regulatory role in Cepharanthine biosynthesis
Abstract
Introduction: Cepharanthine (CEP), a bisbenzylisoquinoline alkaloid (bisBIA) extracted from Stephania japonica, has received significant attention for its anti-coronavirus properties. While ethylene response factors (ERFs) have been reported to regulate the biosynthesis of various alkaloids, their role in regulating CEP biosynthesis remains unexplored.
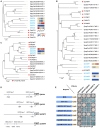
Methods: Genome-wide analysis of the ERF genes was performed with bioinformatics technology, and the expression patterns of different tissues, were analyzed by transcriptome sequencing analysis and real-time quantitative PCR verification. The nuclear-localized ERF gene cluster was shown to directly bind to the promoters of several CEP-associated genes, as demonstrated by yeast one-hybrid assays and subcellular localization assays.
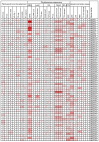
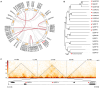
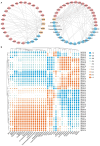
Results: In this work, 59 SjERF genes were identified in the S. japonica genome and further categorized into ten subfamilies. Notably, a SjERF gene cluster containing three SjERF genes was found on chromosome 2. Yeast one-hybrid assays confirmed that the SjERF gene cluster can directly bind to the promoters of several CEP-associated genes, suggesting their crucial role in CEP metabolism. The SjERFs cluster-YFP fusion proteins were observed exclusively in the nuclei of Nicotiana benthamiana leaves. Tissue expression profiling revealed that 13 SjERFs exhibit high expression levels in the root, and the qRT-PCR results of six SjERFs were consistent with the RNA-Seq data. Furthermore, a co-expression network analysis demonstrated that 24 SjERFs were highly positively correlated with the contents of various alkaloids and expression levels of CEP biosynthetic genes.
Conclusion: This study provides the first systematic identification and analysis of ERF transcription factors in the S.japonica genome, laying the foundation for the future functional research of SjERFs transcription factors.
Keywords: Cepharanthine biosynthesis; ERF; Stephania japonica; expression patterns; genome-wide analysis.
Copyright © 2024 Yang, Liu, Ding, Liu, Li, He, Wu, Zhang, Wang, Leng, Chen and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Al-Amin M. Y., Lahiry A., Ferdous R., Hasan M. K., Kader M. A., Alam A. K., et al. (2022). Stephania japonica ameliorates scopolamine-induced memory impairment in mice through inhibition of acetylcholinesterase and oxidative stress. Adv. Pharmacol. Pharm. Sci. 2022, 8305271. doi: 10.1155/2022/8305271 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

