Gasdermin C promotes Stemness and Immune Evasion in Pancreatic Cancer via Pyroptosis-Independent Mechanism
- PMID: 39297408
- PMCID: PMC11558074
- DOI: 10.1002/advs.202308990
Gasdermin C promotes Stemness and Immune Evasion in Pancreatic Cancer via Pyroptosis-Independent Mechanism
Abstract
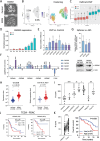
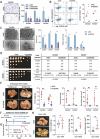
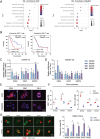
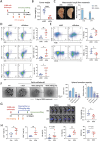
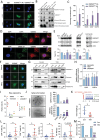
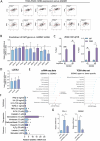
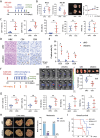
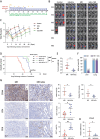
Pancreatic ductal adenocarcinoma (PDAC) is a highly metastatic and lethal disease. Gasdermins are primarily associated with necrosis via membrane permeabilization and pyroptosis, a lytic pro-inflammatory type of cell death. In this study, GSDMC upregulation during PDAC progression is reported. GSDMC directly induces genes related to stemness, EMT, and immune evasion. Targeting Gsdmc in murine PDAC models reprograms the immunosuppressive tumor microenvironment, rescuing the recruitment of anti-tumor immune cells through CXCL9. This not only results in diminished tumor initiation, growth and metastasis, but also enhances the response to KRASG12D inhibition and PD-1 checkpoint blockade, respectively. Mechanistically, it is discovered that ADAM17 cleaves GSDMC, releasing nuclear fragments binding to promoter regions of stemness, metastasis, and immune evasion-related genes. Pharmacological inhibition of GSDMC cleavage or prevention of its nuclear translocation is equally effective in suppressing GSDMC's downstream targets and inhibiting PDAC progression. The findings establish GSDMC as a potential therapeutic target for enhancing treatment response in this deadly disease.
Keywords: KRAS inhibition; cancer stem cells; gasdermin C; immune evasion; immunotherapy; invasion; metastasis; pancreatic ductal adenocarcinoma.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J. Clin. 2021, 71, 209. - PubMed
-
- G. P. C. Collaborators , Lancet Gastroenterol. Hepatol. 2017, 4, 934.
-
- Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J.‐L., Gourgou‐Bourgade S., de la Fouchardière C., Bennouna J., Bachet J.‐B., Khemissa‐Akouz F., Péré‐Vergé D., Delbaldo C., Assenat E., Chauffert B., Michel P., Montoto‐Grillot C., Ducreux M., N. Engl. J. Med. 2011, 364, 1817. - PubMed
-
- Siegel R. L., Miller K. D., Jemal A., CA Cancer J. Clin. 2019, 69, 7. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
