Novel systems biology experimental pipeline reveals matairesinol's antimetastatic potential in prostate cancer: an integrated approach of network pharmacology, bioinformatics, and experimental validation
- PMID: 39297880
- PMCID: PMC11411774
- DOI: 10.1093/bib/bbae466
Novel systems biology experimental pipeline reveals matairesinol's antimetastatic potential in prostate cancer: an integrated approach of network pharmacology, bioinformatics, and experimental validation
Abstract
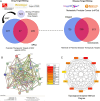
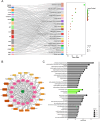
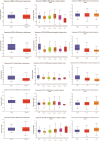
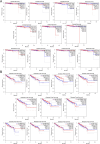
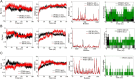
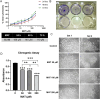
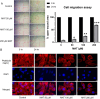
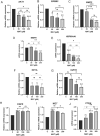
Matairesinol (MAT), a plant lignan renowned for its anticancer properties in hormone-sensitive cancers like breast and prostate cancers, presents a promising yet underexplored avenue in the treatment of metastatic prostate cancer (mPC). To elucidate its specific therapeutic targets and mechanisms, our study adopted an integrative approach, amalgamating network pharmacology (NP), bioinformatics, GeneMANIA-based functional association (GMFA), and experimental validation. By mining online databases, we identified 27 common targets of mPC and MAT, constructing a MAT-mPC protein-protein interaction network via STRING and pinpointing 11 hub targets such as EGFR, AKT1, ERBB2, MET, IGF1, CASP3, HSP90AA1, HIF1A, MMP2, HGF, and MMP9 with CytoHuba. Utilizing DAVID, Gene Ontology (GO) analysis highlighted metastasis-related processes such as epithelial-mesenchymal transition, positive regulation of cell migration, and key Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, including cancer, prostate cancer, PI3K-Akt, and MAPK signaling, while the web resources such as UALCAN and GEPIA2 affirmed the clinical significance of the top 11 hub targets in mPC patient survival analysis and gene expression patterns. Our innovative GMFA enrichment method further enriched network pharmacology findings. Molecular docking analyses demonstrated substantial interactions between MAT and 11 hub targets. Simulation studies confirmed the stable interactions of MAT with selected targets. Experimental validation in PC3 cells, employing quantitative real-time reverse-transcription PCR and various cell-based assays, corroborated MAT's antimetastatic effects on mPC. Thus, this exhaustive NP analysis, complemented by GMFA, molecular docking, molecular dynamics simulations, and experimental validations, underscores MAT's multifaceted role in targeting mPC through diverse therapeutic avenues. Nevertheless, comprehensive in vitro validation is imperative to solidify these findings.
Keywords: PC3 cells; matairesinol; metastatic prostate cancer (mPC); molecular docking; molecular dynamics; network pharmacology.
© The Author(s) 2024. Published by Oxford University Press.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

