SMARCA5-mediated chromatin remodeling is required for germinal center formation
- PMID: 39297882
- PMCID: PMC11413417
- DOI: 10.1084/jem.20240433
SMARCA5-mediated chromatin remodeling is required for germinal center formation
Abstract
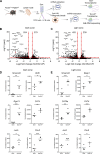
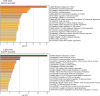
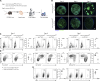
The establishment of long-lasting immunity against pathogens is facilitated by the germinal center (GC) reaction, during which B cells increase their antibody affinity and differentiate into antibody-secreting cells (ASC) and memory cells. These events involve modifications in chromatin packaging that orchestrate the profound restructuring of gene expression networks that determine cell fate. While several chromatin remodelers were implicated in lymphocyte functions, less is known about SMARCA5. Here, using ribosomal pull-down for analyzing translated genes in GC B cells, coupled with functional experiments in mice, we identified SMARCA5 as a key chromatin remodeler in B cells. While the naive B cell compartment remained unaffected following conditional depletion of Smarca5, effective proliferation during B cell activation, immunoglobulin class switching, and as a result GC formation and ASC differentiation were impaired. Single-cell multiomic sequencing analyses revealed that SMARCA5 is crucial for facilitating the transcriptional modifications and genomic accessibility of genes that support B cell activation and differentiation. These findings offer novel insights into the functions of SMARCA5, which can be targeted in various human pathologies.
© 2024 Stoler-Barak et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







References
-
- Alvarez-Saavedra, M., De Repentigny Y., Lagali P.S., Raghu Ram E.V.S., Yan K., Hashem E., Ivanochko D., Huh M.S., Yang D., Mears A.J., et al. . 2014. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat. Commun. 5:4181. 10.1038/ncomms5181 - DOI - PMC - PubMed
-
- Arends, T., Dege C., Bortnick A., Danhorn T., Knapp J.R., Jia H., Harmacek L., Fleenor C.J., Straign D., Walton K., et al. . 2019. CHD4 is essential for transcriptional repression and lineage progression in B lymphopoiesis. Proc. Natl. Acad. Sci. USA. 116:10927–10936. 10.1073/pnas.1821301116 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

