Intestinal immunity in C. elegans is activated by pathogen effector-triggered aggregation of the guard protein TIR-1 on lysosome-related organelles
- PMID: 39299238
- PMCID: PMC11464196
- DOI: 10.1016/j.immuni.2024.08.013
Intestinal immunity in C. elegans is activated by pathogen effector-triggered aggregation of the guard protein TIR-1 on lysosome-related organelles
Abstract
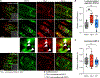
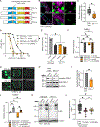
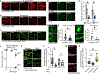
Toll/interleukin-1/resistance (TIR)-domain proteins with enzymatic activity are essential for immunity in plants, animals, and bacteria. However, it is not known how these proteins function in pathogen sensing in animals. We discovered that the lone enzymatic TIR-domain protein in the nematode C. elegans (TIR-1, homolog of mammalian sterile alpha and TIR motif-containing 1 [SARM1]) was strategically expressed on the membranes of a specific intracellular compartment called lysosome-related organelles. The positioning of TIR-1 on lysosome-related organelles enables intestinal epithelial cells in the nematode C. elegans to survey for pathogen effector-triggered host damage. A virulence effector secreted by the bacterial pathogen Pseudomonas aeruginosa alkalinized and condensed lysosome-related organelles. This pathogen-induced morphological change in lysosome-related organelles triggered TIR-1 multimerization, which engaged its intrinsic NAD+ hydrolase (NADase) activity to activate the p38 innate immune pathway and protect the host against microbial intoxication. Thus, TIR-1 is a guard protein in an effector-triggered immune response, which enables intestinal epithelial cells to survey for pathogen-induced host damage.
Keywords: Caenorhabditis elegans; Pseudomonas aeruginosa; SARM1; TIR-1; effector-triggered immunity; intestinal immunity; lysosome-related organelles; phenazines.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Update of
-
Activation of intestinal immunity by pathogen effector-triggered aggregation of lysosomal TIR-1/SARM1.bioRxiv [Preprint]. 2024 Feb 17:2023.12.04.569946. doi: 10.1101/2023.12.04.569946. bioRxiv. 2024. Update in: Immunity. 2024 Oct 8;57(10):2280-2295.e6. doi: 10.1016/j.immuni.2024.08.013. PMID: 38106043 Free PMC article. Updated. Preprint.
References
-
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Osborne Nishimura E, DiAntonio A, Milbrandt J, Dangl JL, and Nishimura MT (2019). TIR domains of plant immune receptors are NAD(+)-cleaving enzymes that promote cell death. Science 365, 799–803. 10.1126/science.aax1771. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

