Prion protein pathology in Ubiquilin 2 models of ALS
- PMID: 39299489
- PMCID: PMC11651290
- DOI: 10.1016/j.nbd.2024.106674
Prion protein pathology in Ubiquilin 2 models of ALS
Abstract
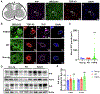
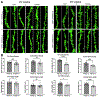
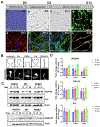
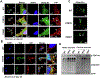
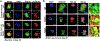
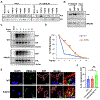
Mutations in UBQLN2 cause ALS and frontotemporal dementia (FTD). The pathological signature in UBQLN2 cases is deposition of highly unusual types of inclusions in the brain and spinal cord that stain positive for UBQLN2. However, what role these inclusions play in pathogenesis remains unclear. Here we show cellular prion protein (PrPC) is found in UBQLN2 inclusions in both mouse and human neuronal induced pluripotent (IPSC) models of UBQLN2 mutations, evidenced by the presence of aggregated forms of PrPC with UBQLN2 inclusions. Turnover studies indicated that the P497H UBQLN2 mutation slows PrPC protein degradation and leads to mislocalization of PrPC in the cytoplasm. Immunoprecipitation studies indicated UBQLN2 and PrPC bind together in a complex. The abnormalities in PrPC caused by UBQLN2 mutations may be relevant in disease pathogenesis.
Keywords: Amyotrophic lateral sclerosis; Human neuronal induced pluripotent models; Mouse models; Prion protein; Ubiquilin 2.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

