PRMT1 inhibition perturbs RNA metabolism and induces DNA damage in clear cell renal cell carcinoma
- PMID: 39300069
- PMCID: PMC11413393
- DOI: 10.1038/s41467-024-52507-y
PRMT1 inhibition perturbs RNA metabolism and induces DNA damage in clear cell renal cell carcinoma
Abstract
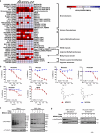
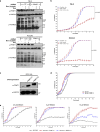
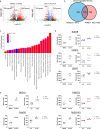
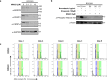
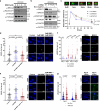
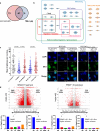
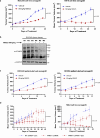
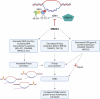
In addition to the ubiquitous loss of the VHL gene in clear cell renal cell carcinoma (ccRCC), co-deletions of chromatin-regulating genes are common drivers of tumorigenesis, suggesting potential vulnerability to epigenetic manipulation. A library of chemical probes targeting a spectrum of epigenetic regulators is screened using a panel of ccRCC models. MS023, a type I protein arginine methyltransferase (PRMT) inhibitor, is identified as an antitumorigenic agent. Individual knockdowns indicate PRMT1 as the specific critical dependency for cancer growth. Further analyses demonstrate impairments to cell cycle and DNA damage repair pathways upon MS023 treatment or PRMT1 knockdown. PRMT1-specific proteomics reveals an interactome rich in RNA binding proteins and further investigation indicates significant widespread disruptions in mRNA metabolism with both MS023 treatment and PRMT1 knockdown, resulting in R-loop accumulation and DNA damage over time. Our data supports PRMT1 as a target in ccRCC and informs a mechanism-based strategy for translational development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Motzer, R. J. et al. Kidney Cancer, Version 3.2015. J. Natl Compr. Cancer Netw.13, 151–159 (2015). - PubMed
-
- Gupta, K., Miller, J. D., Li, J. Z., Russell, M. W. & Charbonneau, C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat. Rev.34, 193–205 (2008). - PubMed
-
- Speed, J. M., Trinh, Q.-D. D., Choueiri, T. K. & Sun, M. Recurrence in localized renal cell carcinoma: a systematic review of contemporary data. Curr. Urol. Rep.18, 15 (2017). - PubMed
-
- Siegel, R. L., Miller, K. D., Jemal, A. & Society, A. C. Cancer statistics, 2020. Ca. Cancer J. Clin.70, 7–30 (2020). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

