Shared neutrophil and T cell dysfunction is accompanied by a distinct interferon signature during severe febrile illnesses in children
- PMID: 39300098
- PMCID: PMC11413185
- DOI: 10.1038/s41467-024-52246-0
Shared neutrophil and T cell dysfunction is accompanied by a distinct interferon signature during severe febrile illnesses in children
Abstract
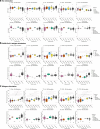
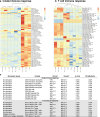
Severe febrile illnesses in children encompass life-threatening organ dysfunction caused by diverse pathogens and other severe inflammatory syndromes. A comparative approach to these illnesses may identify shared and distinct features of host immune dysfunction amenable to immunomodulation. Here, using immunophenotyping with mass cytometry and cell stimulation experiments, we illustrate trajectories of immune dysfunction in 74 children with multi-system inflammatory syndrome in children (MIS-C) associated with SARS-CoV-2, 30 with bacterial infection, 16 with viral infection, 8 with Kawasaki disease, and 42 controls. We explore these findings in a secondary cohort of 500 children with these illnesses and 134 controls. We show that neutrophil activation and apoptosis are prominent in multi-system inflammatory syndrome, and that this is partially shared with bacterial infection. We show that memory T cells from patients with multi-system inflammatory syndrome and bacterial infection are exhausted. In contrast, we show viral infection to be characterized by a distinct signature of decreased interferon signaling and lower interferon receptor gene expression. Improved understanding of immune dysfunction may improve approaches to immunomodulator therapy in severe febrile illnesses in children.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lamontagne, F. et al. A living WHO guideline on drugs for covid-19. BMJ370, m3379 (2020). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- ACL 2018/DH | National Institute for Health Research (NIHR)
- 2016-16-011/DH | National Institute for Health Research (NIHR)
- MRF-160-0008-ELP-KAFO-C0801/Wellcome Trust (Wellcome)
- WT_/Wellcome Trust/United Kingdom
- 848196, 668303, 279185/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

