SPRR1B+ keratinocytes prime oral mucosa for rapid wound healing via STAT3 activation
- PMID: 39300285
- PMCID: PMC11413210
- DOI: 10.1038/s42003-024-06864-5
SPRR1B+ keratinocytes prime oral mucosa for rapid wound healing via STAT3 activation
Abstract
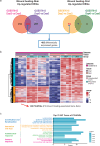
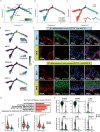
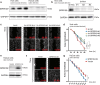
Oral mucosal wounds exhibit accelerated healing with reduced scarring compared to cutaneous wounds, representing an optimal wound healing paradigm. However, the specific cellular subtypes orchestrating the efficient healing of mucosal tissues remain elusive. Through a comprehensive analysis integrating bulk-mRNA and single-cell sequencing data during the wound healing process in oral mucosa and skin, we have delineated a distinct set of genes markedly upregulated during tissue repair. This collection of wound healing-associated genesets was highly enriched in a specific keratinocyte subpopulation identified as STAT3-activated SPRR1B+ keratinocytes. Notably, despite the inherent rapidity of oral mucosal healing, the induction of SPRR1B+ keratinocytes is evident in both skin and mucosal wound healing processes in murine model. Intriguingly, these wound healing-promoting SPRR1B+ keratinocytes, which are induced via STAT3 activation, inherently abundant in unwounded normal mucosa but absent in normal skin. SPRR1B knockdown significantly inhibits mucosal keratinocyte migration, a critical attribute for effective wound healing. In summary, through analysis of human oral and skin wound healing processes at single-cell resolution, coupled with validation in murine model, suggests STAT3-activated SPRR1B+ keratinocytes are associated with the rapid mucosal repair process. This discovery underscores the potential application of SPRR1B+ keratinocytes in the therapeutic management of chronic or non-healing wounds.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

