Expression and regulation of the CXCL9-11 chemokines and CXCR3 receptor in Atlantic salmon (Salmo salar)
- PMID: 39301034
- PMCID: PMC11410577
- DOI: 10.3389/fimmu.2024.1455457
Expression and regulation of the CXCL9-11 chemokines and CXCR3 receptor in Atlantic salmon (Salmo salar)
Abstract
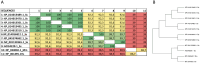
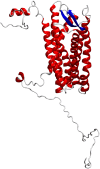
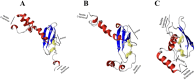
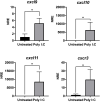
Chemokines are cytokines that mediate leukocyte traffic between the lymphoid organs, the bloodstream, and the site of tissue damage, which is essential for an efficient immune response. In particular, the gamma interferon (IFN- γ) inducible chemokines CXCL9, CXCL10, and CXCL11, and their receptor CXCR3, are involved in T cell and macrophage recruitment to the site of infection. The nature and function of these chemokines and their receptor are well-known in mammals, but further research is needed to achieve a similar level of understanding in fish immunity. Thus, in this study, we seek to identify the genes encoding the components of the Atlantic salmon (Salmo salar) CXCL9, CXCL10, CXCL11/CXCR3 axis (CXCL9-11/CXCR3), predict the protein structure from the amino acid sequence, and explore the regulation of gene expression as well as the response of these chemokines and their receptor to viral infections. The cxcl9, cxcl10, cxcl11, and cxcr3 gene sequences were retrieved from the databases, and the phylogenetic analysis was conducted to determine the evolutionary relationships. The study revealed an interesting pattern of clustering and conservation among fish and mammalian species. The salmon chemokine sequences clustered with orthologs from other fish species, while the mammalian sequences formed separate clades. This indicates a divergent evolution of chemokines between mammals and fish, possibly due to different evolutionary pressures. While the structural analysis of the chemokines and the CXCR3 receptor showed the conservation of critical motifs and domains, suggesting preserved functions and stability throughout evolution. Regarding the regulation of gene expression, some components of the CXCL9-11/CXCR3 axis are induced by recombinant gamma interferon (rIFN-γ) and by Infectious pancreatic necrosis virus (IPNV) infection in Atlantic salmon cells. Further studies are needed to explore the role of Atlantic salmon CXCL9-11 chemokines in regulating immune cell migration and endothelial activation, as seen in mammals. To the best of our knowledge, there have been no functional studies of chemokines to understand these effects in Atlantic salmon.
Keywords: CXCL10; CXCL11; CXCL9; CXCR3; Salmo salar; chemokine; fish immunity; teleost.
Copyright © 2024 Valdés, Espinoza, Pareja-Barrueto, Olate, Barraza-Rojas, Benavides-Larenas, Cortés and Imarai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International union of pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. (2014) 66:1–79. doi: 10.1124/pr.113.007724 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

