Subset-specific mitochondrial stress and DNA damage shape T cell responses to fever and inflammation
- PMID: 39303018
- PMCID: PMC11607909
- DOI: 10.1126/sciimmunol.adp3475
Subset-specific mitochondrial stress and DNA damage shape T cell responses to fever and inflammation
Abstract
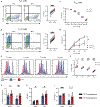
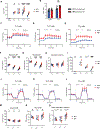
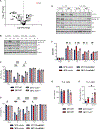
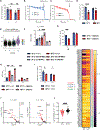
Heat is a cardinal feature of inflammation, yet its impacts on immune cells remain uncertain. We show that moderate-grade fever temperatures (39°C) increased murine CD4 T cell metabolism, proliferation, and inflammatory effector activity while decreasing regulatory T cell suppressive capacity. However, heat-exposed T helper 1 (TH1) cells selectively developed mitochondrial stress and DNA damage that activated Trp53 and stimulator of interferon genes pathways. Although many TH1 cells subjected to such temperatures died, surviving TH1 cells exhibited increased mitochondrial mass and enhanced activity. Electron transport chain complex 1 (ETC1) was rapidly impaired under fever-range temperatures, a phenomenon that was specifically detrimental to TH1 cells. TH1 cells with elevated DNA damage and ETC1 signatures were also detected in human chronic inflammation. Thus, fever-relevant temperatures disrupt ETC1 to selectively drive apoptosis or adaptation of TH1 cells to maintain genomic integrity and enhance effector functions.
Conflict of interest statement
Figures



References
-
- Daniel RM, Danson MJ, Eisenthal R, Lee CK, Peterson ME, The effect of temperature on enzyme activity: New insights and their implications. Extremophiles 12, 51–59 (2008). - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AI153167/AI/NIAID NIH HHS/United States
- U2C DK059637/DK/NIDDK NIH HHS/United States
- R01 CA245134/CA/NCI NIH HHS/United States
- T32 AI112541/AI/NIAID NIH HHS/United States
- T32 DK101003/DK/NIDDK NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- T32 AR059039/AR/NIAMS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 HL136664/HL/NHLBI NIH HHS/United States
- R01 CA217987/CA/NCI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 HL118979/HL/NHLBI NIH HHS/United States
- K00 CA253718/CA/NCI NIH HHS/United States
- U01 AI176244/AI/NIAID NIH HHS/United States
- R01 DK105550/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

