Fusobacterium nucleatum extracellular vesicles are enriched in colorectal cancer and facilitate bacterial adhesion
- PMID: 39303027
- PMCID: PMC11414721
- DOI: 10.1126/sciadv.ado0016
Fusobacterium nucleatum extracellular vesicles are enriched in colorectal cancer and facilitate bacterial adhesion
Abstract
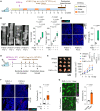
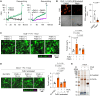
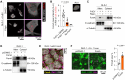
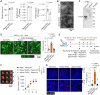
Fusobacterium nucleatum in colorectal cancer (CRC) tissue is implicated at multiple stages of the disease, while the mechanisms underlying bacterial translocation and colonization remain incompletely understood. Herein, we investigated whether extracellular vesicles derived from F. nucleatum (FnEVs) have impacts on bacterial colonization. In mice with colitis-related CRC, a notable enrichment of FnEVs was observed, leading to a significant increase in intratumor colonization by F. nucleatum and accelerated progression of CRC. The enrichment of FnEVs in clinical CRC tissues was demonstrated. Subsequently, we revealed that FnEVs undergo membrane fusion with CRC cells, leading to the transfer and retention of FomA on recipient cell surfaces. Given its ability to facilitate F. nucleatum autoaggregation through interaction with FN1441, the presence of FomA on CRC cell surfaces presents a target for bacterial adhesion. Collectively, the findings unveil a mechanism used by EVs to prepare a niche conducive for bacterial colonization in distal organs.
Figures


References
-
- Wong C. C., Yu J., Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 20, 429–452 (2023). - PubMed
-
- Wang N., Fang J. Y., Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31, 159–172 (2023). - PubMed
-
- Jiang S. S., Xie Y.-L., Xiao X.-Y., Kang Z.-R., Lin X.-L., Zhang L., Li C.-S., Qian Y., Xu P.-P., Leng X.-X., Wang L.-W., Tu S.-P., Zhong M., Zhao G., Chen J.-X., Wang Z., Liu Q., Hong J., Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe 31, 781–e789 (2023). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

