Structural insights into polyamine spermidine uptake by the ABC transporter PotD-PotABC
- PMID: 39303029
- PMCID: PMC11414716
- DOI: 10.1126/sciadv.ado8107
Structural insights into polyamine spermidine uptake by the ABC transporter PotD-PotABC
Abstract
Polyamines, characterized by their polycationic nature, are ubiquitously present in all organisms and play numerous cellular functions. Among polyamines, spermidine stands out as the predominant type in both prokaryotic and eukaryotic cells. The PotD-PotABC protein complex in Escherichia coli, belonging to the adenosine triphosphate-binding cassette transporter family, is a spermidine-preferential uptake system. Here, we report structural details of the polyamine uptake system PotD-PotABC in various states. Our analyses reveal distinct "inward-facing" and "outward-facing" conformations of the PotD-PotABC transporter, as well as conformational changes in the "gating" residues (F222, Y223, D226, and K241 in PotB; Y219 and K223 in PotC) controlling spermidine uptake. Therefore, our structural analysis provides insights into how the PotD-PotABC importer recognizes the substrate-binding protein PotD and elucidates molecular insights into the spermidine uptake mechanism of bacteria.
Figures
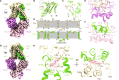

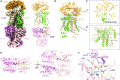



References
-
- Igarashi K., Kashiwagi K., Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51 (2010). - PubMed
-
- Wu D., Lim S. C., Dong Y., Wu J., Tao F., Zhou L., Zhang L. H., Song H., Structural basis of substrate binding specificity revealed by the crystal structures of polyamine receptors SpuD and SpuE from Pseudomonas aeruginosa. J. Mol. Biol. 416, 697–712 (2012). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

