Generation and characterization of giant panda induced pluripotent stem cells
- PMID: 39303041
- PMCID: PMC11414735
- DOI: 10.1126/sciadv.adn7724
Generation and characterization of giant panda induced pluripotent stem cells
Abstract
The giant panda (Ailuropoda melanoleuca) stands as a flagship and umbrella species, symbolizing global biodiversity. While traditional assisted reproductive technology faces constraints in safeguarding the genetic diversity of giant pandas, induced pluripotent stem cells (iPSCs) known for their capacity to differentiate into diverse cells types, including germ cells, present a transformative potential for conservation of endangered animals. In this study, primary fibroblast cells were isolated from the giant panda, and giant panda iPSCs (GPiPSCs) were generated using a non-integrating episomal vector reprogramming method. Characterization of these GPiPSCs revealed their state of primed pluripotency and demonstrated their potential for differentiation. Furthermore, we innovatively formulated a species-specific chemically defined FACL medium and unraveled the intricate signaling pathway networks responsible for maintaining the pluripotency and fostering cell proliferation of GPiPSCs. This study provides key insights into rare species iPSCs, offering materials for panda characteristics research and laying the groundwork for in vitro giant panda gamete generation, potentially aiding endangered species conservation.
Figures
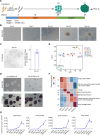
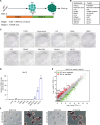





References
-
- Wei F., Fan H., Hu Y., Ailuropoda melanoleuca (Giant Panda). Trends Genet. 36, 68–69 (2020). - PubMed
-
- Kang D., A review of the impacts of four identified major human disturbances on the habitat and habitat use of wild giant pandas from 2015 to 2020. Sci. Total Environ. 763, 142975 (2021). - PubMed
-
- Swaisgood R. R., Wang D., Wei F., Panda downlisted but not out of the woods. Conserv. Lett. 11, e12355 (2018).
-
- Kong L., Xu W., Xiao Y., Pimm S. L., Shi H., Ouyang Z., Spatial models of giant pandas under current and future conditions reveal extinction risks. Nat. Ecol. Evol. 5, 1309–1316 (2021). - PubMed
-
- Zhang M., Hou R., Zheng H., Zhu Q., Zhang Z., Liu Y., Establishment and cryopreservation of a giant panda skin fibroblast cell line. Chin. J. Biol. 40, 61–67 (2005).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

