CheB localizes to polar receptor arrays during repellent adaptation
- PMID: 39303042
- PMCID: PMC11414734
- DOI: 10.1126/sciadv.adp5636
CheB localizes to polar receptor arrays during repellent adaptation
Abstract
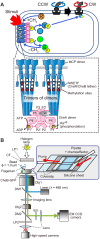
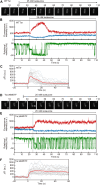
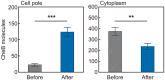
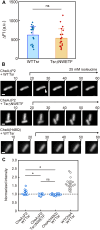
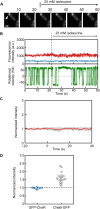
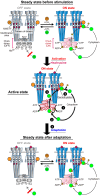
Adaptation of the response to stimuli is a fundamental process for all organisms. Here, we show that the adaptation enzyme CheB methylesterase of Escherichia coli assembles to the ON state receptor array after exposure to the repellent l-isoleucine and dissociates from the array after adaptation is complete. The duration of increased CheB localization and the time of highly clockwise-biased flagellar rotation were similar and depended on the strength of the stimulus. The increase in CheB at the receptor array and the decrease in cytoplasmic CheB were both ~100 molecules, which represents 15 to 20% of the total cellular content of CheB. We confirmed that the main binding site for CheB in the ON state array is the P2 domain of phosphorylated CheA, with a second minor site being the carboxyl-terminal pentapeptide of the serine chemoreceptor. Thus, we have been able to quantify the regulation of the signal output of the receptor array by the intracellular dynamics of an adaptation enzyme.
Figures

References
-
- Wadhams G. H., Armitage J. P., Making sense of it all: Bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037 (2004). - PubMed
-
- R. Macnab, “Flagela and motility” in Eschericia coli and Salmonella, F. C. Neidhardt, Ed. (American Society for Microbiology, 1996), pp.123–145.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

