Spatial analysis reveals targetable macrophage-mediated mechanisms of immune evasion in hepatocellular carcinoma minimal residual disease
- PMID: 39304772
- PMCID: PMC12258040
- DOI: 10.1038/s43018-024-00828-8
Spatial analysis reveals targetable macrophage-mediated mechanisms of immune evasion in hepatocellular carcinoma minimal residual disease
Abstract
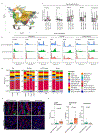
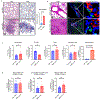
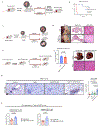
Hepatocellular carcinoma (HCC) frequently recurs from minimal residual disease (MRD), which persists after therapy. Here, we identified mechanisms of persistence of residual tumor cells using post-chemoembolization human HCC (n = 108 patients, 1.07 million cells) and a transgenic mouse model of MRD. Through single-cell high-plex cytometric imaging, we identified a spatial neighborhood within which PD-L1 + M2-like macrophages interact with stem-like tumor cells, correlating with CD8+ T cell exhaustion and poor survival. Further, through spatial transcriptomics of residual HCC, we showed that macrophage-derived TGFβ1 mediates the persistence of stem-like tumor cells. Last, we demonstrate that combined blockade of Pdl1 and Tgfβ excluded immunosuppressive macrophages, recruited activated CD8+ T cells and eliminated residual stem-like tumor cells in two mouse models: a transgenic model of MRD and a syngeneic orthotopic model of doxorubicin-resistant HCC. Thus, our spatial analyses reveal that PD-L1+ macrophages sustain MRD by activating the TGFβ pathway in stem-like cancer cells and targeting this interaction may prevent HCC recurrence from MRD.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
A.T.M., A.E.T., R.P. and H.B.D. are employees of Enable Medicine. All other authors declare no competing interests.
Figures















References
-
- Kim DJ et al. Recurrence of hepatocellular carcinoma: importance of mRECIST response to chemoembolization and tumor size. Am. J. Transplant 14, 1383–1390 (2014). - PubMed
-
- Agopian VG et al. Impact of pretransplant bridging locoregional therapy for patients with hepatocellular carcinoma within Milan criteria undergoing liver transplantation: analysis of 3601 patients from the US multicenter HCC transplant consortium. Ann. Surg 266, 525–535 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- R35 CA253180/CA/NCI NIH HHS/United States
- CA208735/Center for Strategic Scientific Initiatives, National Cancer Institute (NCI Center for Strategic Scientific Initiatives)
- R01 CA208735/CA/NCI NIH HHS/United States
- CA253180/Center for Strategic Scientific Initiatives, National Cancer Institute (NCI Center for Strategic Scientific Initiatives)
- K08 CA222676/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

