TDP-43 regulates LC3ylation in neural tissue through ATG4B cryptic splicing inhibition
- PMID: 39305312
- PMCID: PMC11416411
- DOI: 10.1007/s00401-024-02780-4
TDP-43 regulates LC3ylation in neural tissue through ATG4B cryptic splicing inhibition
Abstract
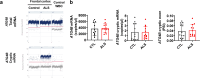
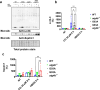
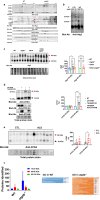
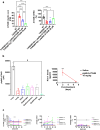
Amyotrophic lateral sclerosis (ALS) is an adult-onset motor neuron disease with a mean survival time of three years. The 97% of the cases have TDP-43 nuclear depletion and cytoplasmic aggregation in motor neurons. TDP-43 prevents non-conserved cryptic exon splicing in certain genes, maintaining transcript stability, including ATG4B, which is crucial for autophagosome maturation and Microtubule-associated proteins 1A/1B light chain 3B (LC3B) homeostasis. In ALS mice (G93A), Atg4b depletion worsens survival rates and autophagy function. For the first time, we observed an elevation of LC3ylation in the CNS of both ALS patients and atg4b-/- mouse spinal cords. Furthermore, LC3ylation modulates the distribution of ATG3 across membrane compartments. Antisense oligonucleotides (ASOs) targeting cryptic exon restore ATG4B mRNA in TARDBP knockdown cells. We further developed multi-target ASOs targeting TDP-43 binding sequences for a broader effect. Importantly, our ASO based in peptide-PMO conjugates show brain distribution post-IV administration, offering a non-invasive ASO-based treatment avenue for neurodegenerative diseases.
Keywords: ALS; Antisense oligonucleotides; Autophagy; Digital PCR; Post-translational modification.
© 2024. The Author(s).
Conflict of interest statement
P.T., M.J.A.W., M.A.V., M.P.-O. are inventors in a patent application (2315883.5), priority request in UK covering the use of ASOs targeting ATG4B cryptic exon and multi-targets for TDP-43 proteinopathies.
Figures


References
-
- Burki U, Keane J, Blain A, O’Donovan L, Gait MJ, Laval SH et al (2015) Development and application of an ultrasensitive hybridization-based ELISA method for the determination of peptide-conjugated phosphorodiamidate morpholino oligonucleotides. Nucleic Acid Ther 25:275. 10.1089/NAT.2014.0528 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

