Geranylgeranylated SCFFBXO10 regulates selective outer mitochondrial membrane proteostasis and function
- PMID: 39306844
- PMCID: PMC11573457
- DOI: 10.1016/j.celrep.2024.114783
Geranylgeranylated SCFFBXO10 regulates selective outer mitochondrial membrane proteostasis and function
Abstract
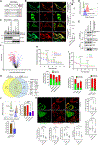
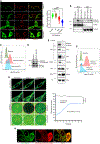
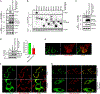
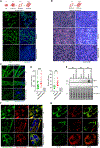
Compartment-specific cellular membrane protein turnover is not well understood. We show that FBXO10, the interchangeable component of the cullin-RING-ligase 1 complex, undergoes lipid modification with geranylgeranyl isoprenoid at cysteine953, facilitating its dynamic trafficking to the outer mitochondrial membrane (OMM). FBXO10 polypeptide lacks a canonical mitochondrial targeting sequence (MTS); instead, its geranylgeranylation at C953 and interaction with two cytosolic factors, cytosolic factor-like δ subunit of type 6 phosphodiesterase (PDE6δ; a prenyl-group-binding protein) and heat shock protein 90 (HSP90; a chaperone), orchestrate specific OMM targeting of prenyl-FBXO10. The FBXO10(C953S) mutant redistributes away from the OMM, impairs mitochondrial ATP production and membrane potential, and increases fragmentation. Phosphoglycerate mutase-5 (PGAM5) was identified as a potential substrate of FBXO10 at the OMM using comparative quantitative proteomics of enriched mitochondria. FBXO10 loss or expression of prenylation-deficient FBXO10(C953S) inhibited PGAM5 degradation, disrupted mitochondrial homeostasis, and impaired myogenic differentiation of human induced pluripotent stem cells (iPSCs) and murine myoblasts. Our studies identify a mechanism for FBXO10-mediated regulation of selective mitochondrial proteostasis potentially amenable to therapeutic intervention.
Keywords: CP: Metabolism; CP: Molecular biology; E3-ligase; F-box protein; FBXO10; HSP90; PDE6δ; mitochondria; prenylation; trafficking; ubiquitination.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Update of
-
Geranylgeranylated-SCFFBXO10 Regulates Selective Outer Mitochondrial Membrane Proteostasis and Function.bioRxiv [Preprint]. 2024 Apr 16:2024.04.16.589745. doi: 10.1101/2024.04.16.589745. bioRxiv. 2024. Update in: Cell Rep. 2024 Oct 22;43(10):114783. doi: 10.1016/j.celrep.2024.114783. PMID: 38659932 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

