Single-cell exome sequencing reveals polyclonal seeding and TRPS1 mutations in colon cancer metastasis
- PMID: 39307879
- PMCID: PMC11417107
- DOI: 10.1038/s41392-024-01960-8
Single-cell exome sequencing reveals polyclonal seeding and TRPS1 mutations in colon cancer metastasis
Abstract
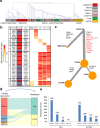
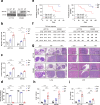
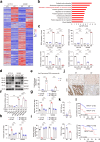
Liver metastasis remains the primary cause of mortality in patients with colon cancer. Identifying specific driver gene mutations that contribute to metastasis may offer viable therapeutic targets. To explore clonal evolution and genetic heterogeneity within the metastasis, we conducted single-cell exome sequencing on 150 single cells isolated from the primary tumor, liver metastasis, and lymphatic metastasis from a stage IV colon cancer patient. The genetic landscape of the tumor samples revealed that both lymphatic and liver metastases originated from the same region of the primary tumor. Notably, the liver metastasis was derived directly from the primary tumor, bypassing the lymph nodes. Comparative analysis of the sequencing data for individual cell pairs within different tumors demonstrated that the genetic heterogeneity of both liver and lymphatic metastases was also greater than that of the primary tumor. This finding indicates that liver and lymphatic metastases arose from clusters of circulating tumor cell (CTC) of a polyclonal origin, rather than from a single cell from the primary tumor. Single-cell transcriptome analysis suggested that higher EMT score and CNV scores were associated with more polyclonal metastasis. Additionally, a mutation in the TRPS1 (Transcriptional repressor GATA binding 1) gene, TRPS1 R544Q, was enriched in the single cells from the liver metastasis. The mutation significantly increased CRC invasion and migration both in vitro and in vivo through the TRPS1R544Q/ZEB1 axis. Further TRPS1 mutations were detected in additional colon cancer cases, correlating with advanced-stage disease and inferior prognosis. These results reveal polyclonal seeding and TRPS1 mutation as potential mechanisms driving the development of liver metastases in colon cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

