A systematic framework for understanding the microbiome in human health and disease: from basic principles to clinical translation
- PMID: 39307902
- PMCID: PMC11418828
- DOI: 10.1038/s41392-024-01946-6
A systematic framework for understanding the microbiome in human health and disease: from basic principles to clinical translation
Abstract
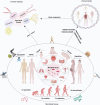
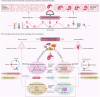
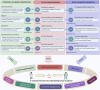
The human microbiome is a complex and dynamic system that plays important roles in human health and disease. However, there remain limitations and theoretical gaps in our current understanding of the intricate relationship between microbes and humans. In this narrative review, we integrate the knowledge and insights from various fields, including anatomy, physiology, immunology, histology, genetics, and evolution, to propose a systematic framework. It introduces key concepts such as the 'innate and adaptive genomes', which enhance genetic and evolutionary comprehension of the human genome. The 'germ-free syndrome' challenges the traditional 'microbes as pathogens' view, advocating for the necessity of microbes for health. The 'slave tissue' concept underscores the symbiotic intricacies between human tissues and their microbial counterparts, highlighting the dynamic health implications of microbial interactions. 'Acquired microbial immunity' positions the microbiome as an adjunct to human immune systems, providing a rationale for probiotic therapies and prudent antibiotic use. The 'homeostatic reprogramming hypothesis' integrates the microbiome into the internal environment theory, potentially explaining the change in homeostatic indicators post-industrialization. The 'cell-microbe co-ecology model' elucidates the symbiotic regulation affecting cellular balance, while the 'meta-host model' broadens the host definition to include symbiotic microbes. The 'health-illness conversion model' encapsulates the innate and adaptive genomes' interplay and dysbiosis patterns. The aim here is to provide a more focused and coherent understanding of microbiome and highlight future research avenues that could lead to a more effective and efficient healthcare system.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

