Spatiotemporal Analysis of Mesenchymal Stem Cells Fate Determination by Inflammatory Niche Following Soft Tissue Injury at a Single-Cell Level
- PMID: 39308190
- PMCID: PMC11578311
- DOI: 10.1002/advs.202310282
Spatiotemporal Analysis of Mesenchymal Stem Cells Fate Determination by Inflammatory Niche Following Soft Tissue Injury at a Single-Cell Level
Abstract
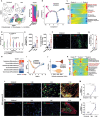
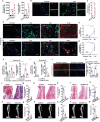
Heterotopic ossification (HO), often arising in response to traumatic challenges, results from the aberrant osteochondral differentiation of mesenchymal stem cells (MSCs). Nevertheless, the impact of trauma-induced inflammatory exposure on MSC fate determination remains ambiguous. In this study, the cellular diversity within inflammatory lesions is elucidated, comprising MSCs and several innate and adaptive immune cells. It is observed that quiescent MSCs transition into cycling MSCs, subsequently giving rise to chondrogenic (cMSC) and/or osteogenic (oMSC) lineages within the inflammatory microenvironment following muscle or tendon injuries, as revealed through single-cell RNA sequencing (scRNA-seq), spatial transcriptome and lineage tracing analysis. Moreover, these investigations demonstrate that neutrophils and natural killer (NK) cells enhance transition of quiescent MSCs into cycling MSCs, which is also controlled by M1 macrophages, a subpopulation of macrophages can also stimulate cMSC and oMSC production from cycling MSCs. Additionally, M2 macrophages, CD4+ and CD8+ T lymphocytes are found to promote chondrogenesis. Further analysis demonstrates that immune cells promotes the activation of signaling transducers and activators of transcription (STAT) pathway and phosphoinositide 3 (PI3K)/protein kinase B (AKT) pathway in MSC proliferation and osteochondral progenitors' production, respectively. These findings highlight the dynamics of MSC fate within the inflammatory lesion and unveil the molecular landscape of osteoimmunological interactions, which holds promise for advancing HO treatment.
Keywords: heterotopic ossification; inflammatory microenvironment; mesenchymal stem cells; osteoimmunology.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Shore E. M., Ahn J., de Beur S. J., Li M., Xu M., Gardner R. J. M., Zasloff M. A., Whyte M. P., Levine M. A., Kaplan F. S., N. Engl. J. Med. 2002, 346, 99. - PubMed
MeSH terms
Grants and funding
- 82173004/Natural Science Foundation of China
- 82102573/Natural Science Foundation of China
- 81870085/Natural Science Foundation of China
- GZC20240010/Postdoctoral Fellowship Program of CPSF
- YJS20210259/Chinese scholarship council Research Collaboration Project, Scientific Research Project
- 2019xkjT004/Chinese scholarship council Research Collaboration Project, Scientific Research Project
- XJ2020019/Chinese scholarship council Research Collaboration Project, Scientific Research Project
- 0107048202/Chinese scholarship council Research Collaboration Project, Scientific Research Project
- GXXT-2021-063/Collaborative Innovation Project of Anhui Universities
LinkOut - more resources
Full Text Sources
Research Materials
