Hypoxia-responsive zinc finger E-box-binding homeobox 2 (ZEB2) regulates a network of calcium-handling genes in the injured heart
- PMID: 39308239
- PMCID: PMC11630050
- DOI: 10.1093/cvr/cvae163
Hypoxia-responsive zinc finger E-box-binding homeobox 2 (ZEB2) regulates a network of calcium-handling genes in the injured heart
Abstract
Aims: Intracellular calcium (Ca2+) overload is known to play a critical role in the development of cardiac dysfunction. Despite the remarkable improvement in managing the progression of heart disease, developing effective therapies for heart failure (HF) remains a challenge. A better understanding of molecular mechanisms that maintain proper Ca2+ levels and contractility in the injured heart could be of therapeutic value.
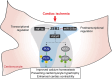
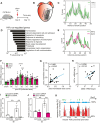
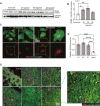
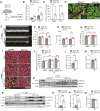
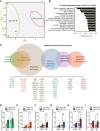
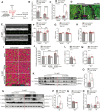
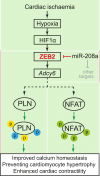
Methods and results: Here, we report that transcription factor zinc finger E-box-binding homeobox 2 (ZEB2) is induced by hypoxia-inducible factor 1-alpha (HIF1α) in hypoxic cardiomyocytes and regulates a network of genes involved in Ca2+ handling and contractility during ischaemic heart disease. Gain- and loss-of-function studies in genetic mouse models revealed that ZEB2 expression in cardiomyocytes is necessary and sufficient to protect the heart against ischaemia-induced diastolic dysfunction and structural remodelling. Moreover, RNA sequencing of ZEB2-overexpressing (Zeb2 cTg) hearts post-injury implicated ZEB2 in regulating numerous Ca2+-handling and contractility-related genes. Mechanistically, ZEB2 overexpression increased the phosphorylation of phospholamban at both serine-16 and threonine-17, implying enhanced activity of sarcoplasmic reticulum Ca2+-ATPase (SERCA2a), thereby augmenting SR Ca2+ uptake and contractility. Furthermore, we observed a decrease in the activity of Ca2+-dependent calcineurin/NFAT signalling in Zeb2 cTg hearts, which is the main driver of pathological cardiac remodelling. On a post-transcriptional level, we showed that ZEB2 expression can be regulated by the cardiomyocyte-specific microRNA-208a (miR-208a). Blocking the function of miR-208a with anti-miR-208a increased ZEB2 expression in the heart and effectively protected from the development of pathological cardiac hypertrophy.
Conclusion: Together, we present ZEB2 as a central regulator of contractility and Ca2+-handling components in the mammalian heart. Further mechanistic understanding of the role of ZEB2 in regulating Ca2+ homeostasis in cardiomyocytes is an essential step towards the development of improved therapies for HF.
Keywords: Calcium handling; Cardiac ischaemia; Hypoxia; MicroRNA; Post-transcriptional regulation; Transcriptional regulation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: none declared.
Figures

References
-
- Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Shay CM, Spartano NL, Stokes A, Tirschwell DL, VanWagner LB, Tsao CW, American Heart Association Council on E, Prevention Statistics C, Stroke Statistics S . Heart Disease and Stroke Statistics-2020 Update: a report from the American Heart Association. Circulation 2020;141:e139–e596. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

