ZNF197-AS1/miR-425/GABARAPL1 axis: a novel regulatory mechanism in uveal melanoma
- PMID: 39308299
- PMCID: PMC11774234
- DOI: 10.1152/ajpcell.00457.2024
ZNF197-AS1/miR-425/GABARAPL1 axis: a novel regulatory mechanism in uveal melanoma
Abstract
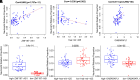
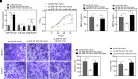
This study investigates the role of the long noncoding RNA (lncRNA) ZNF197-AS1 in uveal melanoma (UM), focusing on its function within a competing endogenous RNA (ceRNA) network. Using the UM-related TCGA (The Cancer Genome Atlas) dataset, we analyzed the expression levels of ZNF197-AS1 and its correlation with miR-425 and GABARAPL1, an essential autophagy-related gene. Our analysis revealed that ZNF197-AS1 acts as a ceRNA by competitively binding to miR-425, resulting in the upregulation of GABARAPL1. This interaction plays a crucial role in the growth and metastasis of UM. The expression of GABARAPL1 showed a strong correlation with the clinical outcomes of patients with UM. Furthermore, in vitro assays confirmed that ZNF197-AS1 impedes UM cell proliferation, migration, and invasion by modulating the miR-425/GABARAPL1 axis. These findings suggest that ZNF197-AS1 can effectively inhibit UM progression through this ceRNA regulatory network. This study provides valuable insights into the molecular mechanisms underlying UM and highlights the potential of targeting the ZNF197-AS1/miR-425/GABARAPL1 axis as a therapeutic strategy for UM.NEW & NOTEWORTHY This study identifies the ZNF197-AS1/miR-425/GABARAPL1 axis as a novel regulatory mechanism in uveal melanoma. ZNF197-AS1 upregulates GABARAPL1 by sponging miR-425, inhibiting UM cell proliferation, migration, and invasion. This discovery highlights a potential therapeutic target, providing new insights into UM progression and patient outcomes.
Keywords: GABARAPL1; ZNF197-AS1; ceRNA regulatory network; miR-425; uveal melanoma.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures







References
-
- Elder DE, Bastian BC, Cree IA, Massi D, Scolyer RA. The 2018 World Health Organization Classification of cutaneous, mucosal, and uveal melanoma: detailed analysis of 9 distinct subtypes defined by their evolutionary pathway. Arch Pathol Lab Med 144: 500–522, 2020. doi: 10.5858/arpa.2019-0561-RA. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

