Aberrant neurodevelopment in human iPS cell-derived models of Alexander disease
- PMID: 39308436
- PMCID: PMC11660530
- DOI: 10.1002/glia.24618
Aberrant neurodevelopment in human iPS cell-derived models of Alexander disease
Abstract
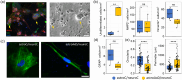
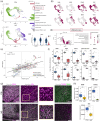
Alexander disease (AxD) is a rare and severe neurodegenerative disorder caused by mutations in glial fibrillary acidic protein (GFAP). While the exact disease mechanism remains unknown, previous studies suggest that mutant GFAP influences many cellular processes, including cytoskeleton stability, mechanosensing, metabolism, and proteasome function. While most studies have primarily focused on GFAP-expressing astrocytes, GFAP is also expressed by radial glia and neural progenitor cells, prompting questions about the impact of GFAP mutations on central nervous system (CNS) development. In this study, we observed impaired differentiation of astrocytes and neurons in co-cultures of astrocytes and neurons, as well as in neural organoids, both generated from AxD patient-derived induced pluripotent stem (iPS) cells with a GFAPR239C mutation. Leveraging single-cell RNA sequencing (scRNA-seq), we identified distinct cell populations and transcriptomic differences between the mutant GFAP cultures and a corrected isogenic control. These findings were supported by results obtained with immunocytochemistry and proteomics. In co-cultures, the GFAPR239C mutation resulted in an increased abundance of immature cells, while in unguided neural organoids and cortical organoids, we observed altered lineage commitment and reduced abundance of astrocytes. Gene expression analysis revealed increased stress susceptibility, cytoskeletal abnormalities, and altered extracellular matrix and cell-cell communication patterns in the AxD cultures, which also exhibited higher cell death after stress. Overall, our results point to altered cell differentiation in AxD patient-derived iPS-cell models, opening new avenues for AxD research.
Keywords: Alexander disease; GFAP; iPS cells; neural organoids.
© 2024 The Author(s). GLIA published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Basak, M. , Sengar, A. S. , Das, K. , Mahata, T. , Kumar, M. , Kumar, D. , Biswas, S. , Sarkar, S. , Kumar, P. , Das, P. , Stewart, A. , & Maity, B. (2023). A RGS7‐CaMKII complex drives myocyte‐intrinsic and myocyte‐extrinsic mechanisms of chemotherapy‐induced cardiotoxicity. Proceedings of the National Academy of Sciences, 120(1), e2213537120. 10.1073/pnas.2213537120 - DOI - PMC - PubMed
-
- Battaglia, R. A. , Beltran, A. S. , Delic, S. , Dumitru, R. , Robinson, J. A. , Kabiraj, P. , Herring, L. E. , Madden, V. J. , Ravinder, N. , Willems, E. , Newman, R. A. , Quinlan, R. A. , Goldman, J. E. , Perng, M. D. , Inagaki, M. , & Snider, N. T. (2019). Site‐specific phosphorylation and caspase cleavage of GFAP are new markers of Alexander disease severity. eLife, 8, e47789. 10.7554/eLife.47789 - DOI - PMC - PubMed
-
- Belvindrah, R. , Graus‐Porta, D. , Goebbels, S. , Nave, K. A. , & Muller, U. (2007). Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. The Journal of Neuroscience, 27(50), 13854–13865. 10.1523/JNEUROSCI.4494-07.2007 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 825575/EJP RD - European Joint Programme on Rare Diseases
- 2017-02255/Swedish Research Council
- 2019-00284/Swedish Research Council
- 2020-01148/Swedish Research Council
- LCF/PR/HR21/52410002/'la Caixa' Foundation
- 146051/Avtal om Läkarutbildning och Forskning (ALF) Gothenburg
- 965939/Avtal om Läkarutbildning och Forskning (ALF) Gothenburg
- Amlöv's Foundation
- PID2021-126827OB-I00/E. Jacobson's Donation Fund
- RVO 86652036/Institutional support (Czech Republic)
- 24-11364S/Czech Science Foundation
- 24-12028S/Czech Science Foundation
- Petrus och Augusta Hedlunds stiftelse
- FO02021-0082/Hjärnfonden
- PID2021-126827OB-I00/MCIN/AEI/10.13039/501100011033/ERDF
- SM23-0033/Swedish Foundation for Strategic Research
- 184.034.019/X-Omics initiative
- Swedish Society for Medical Research
- Söderberg's Foundations
- Hagströmer's Foundation Millennium
- 463002004/ZONMW_/ZonMw/Netherlands
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

