Development of a robust parallel and multi-composite machine learning model for improved diagnosis of Alzheimer's disease: correlation with dementia-associated drug usage and AT(N) protein biomarkers
- PMID: 39308946
- PMCID: PMC11412962
- DOI: 10.3389/fnins.2024.1391465
Development of a robust parallel and multi-composite machine learning model for improved diagnosis of Alzheimer's disease: correlation with dementia-associated drug usage and AT(N) protein biomarkers
Abstract
Introduction: Machine learning (ML) algorithms and statistical modeling offer a potential solution to offset the challenge of diagnosing early Alzheimer's disease (AD) by leveraging multiple data sources and combining information on neuropsychological, genetic, and biomarker indicators. Among others, statistical models are a promising tool to enhance the clinical detection of early AD. In the present study, early AD was diagnosed by taking into account characteristics related to whether or not a patient was taking specific drugs and a significant protein as a predictor of Amyloid-Beta (Aβ), tau, and ptau [AT(N)] levels among participants.
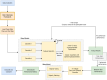
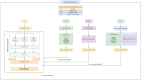
Methods: In this study, the optimization of predictive models for the diagnosis of AD pathologies was carried out using a set of baseline features. The model performance was improved by incorporating additional variables associated with patient drugs and protein biomarkers into the model. The diagnostic group consisted of five categories (cognitively normal, significant subjective memory concern, early mildly cognitively impaired, late mildly cognitively impaired, and AD), resulting in a multinomial classification challenge. In particular, we examined the relationship between AD diagnosis and the use of various drugs (calcium and vitamin D supplements, blood-thinning drugs, cholesterol-lowering drugs, and cognitive drugs). We propose a hybrid-clinical model that runs multiple ML models in parallel and then takes the majority's votes, enhancing the accuracy. We also assessed the significance of three cerebrospinal fluid biomarkers, Aβ, tau, and ptau in the diagnosis of AD. We proposed that a hybrid-clinical model be used to simulate the MRI-based data, with five diagnostic groups of individuals, with further refinement that includes preclinical characteristics of the disorder. The proposed design builds a Meta-Model for four different sets of criteria. The set criteria are as follows: to diagnose from baseline features, baseline and drug features, baseline and protein features, and baseline, drug and protein features.
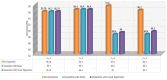
Results: We were able to attain a maximum accuracy of 97.60% for baseline and protein data. We observed that the constructed model functioned effectively when all five drugs were included and when any single drug was used to diagnose the response variable. Interestingly, the constructed Meta-Model worked well when all three protein biomarkers were included, as well as when a single protein biomarker was utilized to diagnose the response variable.
Discussion: It is noteworthy that we aimed to construct a pipeline design that incorporates comprehensive methodologies to detect Alzheimer's over wide-ranging input values and variables in the current study. Thus, the model that we developed could be used by clinicians and medical experts to advance Alzheimer's diagnosis and as a starting point for future research into AD and other neurodegenerative syndromes.
Keywords: Alzheimer's disease; biomarker; drug; early diagnosis; hybrid clinical model; machine learning; multinomial classification; protein.
Copyright © 2024 Khan, Zubair, Shuaib, Sheneamer, Alam and Assiri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aboneh T., Rorissa A., Srinivasagan R. (2022). Stacking-based ensemble learning method for multi-spectral image classification. Technologies 10:17. 10.3390/technologies10010017 - DOI
-
- Aguilera A. M., Escabias M., Valderrama M. J. (2006). Using principal components for estimating logistic regression with high-dimensional multicollinear data. Comput. Stat. Data Anal. 50, 1905–1924. 10.1016/j.csda.2005.03.011 - DOI
-
- Alqahtani N., Alam S., Aqeel I., Shuaib M., Mohsen Khormi I., Khan S. B., et al. (2023). Deep belief networks (DBN) with IoT-based alzheimer's disease detection and classification. Appl. Sci. 13:7833. 10.3390/app13137833 - DOI
LinkOut - more resources
Full Text Sources