Comparative mitogenomic analysis of Sporisorium reilianum f. sp. zeae suggests recombination events during its evolutionary history
- PMID: 39308980
- PMCID: PMC11413489
- DOI: 10.3389/fphys.2024.1264359
Comparative mitogenomic analysis of Sporisorium reilianum f. sp. zeae suggests recombination events during its evolutionary history
Abstract
Introduction: Modern understanding of the concept of genetic diversity must include the study of both nuclear and organellar DNA, which differ greatly in terms of their structure, organization, gene content and distribution. This study comprises an analysis of the genetic diversity of the smut fungus Sporisorium reilianum f. sp. zeae from a mitochondrial perspective.
Methods: Whole-genome sequencing data was generated from biological samples of S. reilianum collected from different geographical regions. Multiple sequence alignment and gene synteny analysis were performed to further characterize genetic diversity in the context of mitogenomic polymorphisms.
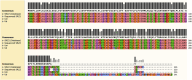
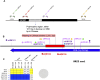
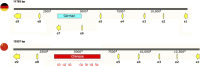
Results: Mitochondria of strains collected in China contained unique sequences. The largest unique sequence stretch encompassed a portion of cox1, a mitochondrial gene encoding one of the subunits that make up complex IV of the mitochondrial electron transport chain. This unique sequence had high percent identity to the mitogenome of the related species Sporisorium scitamineum and Ustilago bromivora.
Discussion: The results of this study hint at potential horizontal gene transfer or mitochondrial genome recombination events during the evolutionary history of basidiomycetes. Additionally, the distinct polymorphic region detected in the Chinese mitogenome provides the ideal foundation to develop a diagnostic method to discern between mitotypes and enhance knowledge on the genetic diversity of this organism.
Keywords: horizontal gene transfer; mitochondria; mitogenome evolution; population mitogenomics; smut fungi.
Copyright © 2024 Mendoza, Lamb, Thomas, Tavares, Schroeder, Müller, Agrawal, Schirawski and Perlin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
LinkOut - more resources
Full Text Sources

