Functional Study of desKR: a Lineage-Specific Two-Component System Positively Regulating Staphylococcus aureus Biofilm Formation
- PMID: 39309069
- PMCID: PMC11416117
- DOI: 10.2147/IDR.S485049
Functional Study of desKR: a Lineage-Specific Two-Component System Positively Regulating Staphylococcus aureus Biofilm Formation
Retraction in
-
Functional Study of desKR: A Lineage-Specific Two-Component System Positively Regulating Staphylococcus Aureus Biofilm Formation [Retraction].Infect Drug Resist. 2024 Dec 13;17:5579-5580. doi: 10.2147/IDR.S509159. eCollection 2024. Infect Drug Resist. 2024. PMID: 39697559 Free PMC article.
Abstract
Purpose: Biofilms significantly contribute to the persistence and antibiotic resistance of Staphylococcus aureus infections. However, the regulatory mechanisms governing biofilm formation of S. aureus remain not fully elucidated. This study aimed to investigate the function of the S. aureus lineage-specific two-component system, desKR, in biofilm regulation and pathogenicity.
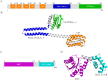
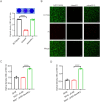
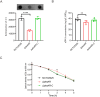
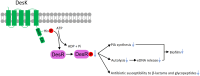
Methods: Bioinformatic analysis was conducted to assess the prevalence of desKR across various S. aureus lineages and to examine its structural features. The impact of desKR on S. aureus pathogenicity was evaluated using in vivo mouse models, including skin abscess, bloodstream infection, and nasal colonization models. Crystal violet staining and confocal laser scanning microscopy were utilized to examine the impact of desKR on S. aureus biofilm formation. Mechanistic insights into desKR-mediated biofilm regulation were investigated by quantifying polysaccharide intercellular adhesin (PIA) production, extracellular DNA (eDNA) release, autolysis assays, and RT-qPCR.
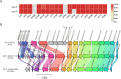
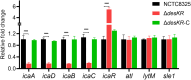
Results: The prevalence of desKR varied among different S. aureus lineages, with notably low carriage rates in ST398 and ST59 lineages. Deletion of desKR in NCTC8325 strain resulted in decreased susceptibility to β-lactam and glycopeptide antibiotics. Although desKR did not significantly affect acute pathogenicity, the ΔdesKR mutant exhibited significantly reduced nasal colonization and biofilm-forming ability. Overexpression of desKR in naturally desKR-lacking strains (ST398 and ST59) enhanced biofilm formation, suggesting a lineage-independent effect. Phenotypic assays further revealed that the ΔdesKR mutant showed reduced PIA production, decreased eDNA release, and lower autolysis rates. RT-qPCR indicated significant downregulation of icaA, icaD, icaB, and icaC genes, along with upregulation of icaR, whereas autolysis-related genes remained unchanged.
Conclusion: The desKR two-component system positively regulates S. aureus biofilm formation in a lineage-independent manner, primarily by modulating PIA synthesis via the ica operon. These findings provide new insights into the molecular mechanisms of biofilm formation in S. aureus and highlight desKR as a potential target for therapeutic strategies aimed at combating biofilm-associated infections.
Keywords: Staphylococcus aureus; biofilm; desKR; two-component system.
© 2024 Ma et al.
Conflict of interest statement
The authors declare no conflicts of interest in this work.
Figures

References
-
- Shree P, Singh CK, Sodhi KK, Surya JN, Singh DK. Biofilms: understanding the structure and contribution towards bacterial resistance in antibiotics. Med Microecol. 2023;16:100084. doi: 10.1016/j.medmic.2023.100084 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

