Dendritic Cell-Related Gene Signatures in Hepatocellular Carcinoma: An Analysis for Prognosis and Therapy Efficacy Evaluation
- PMID: 39309303
- PMCID: PMC11416124
- DOI: 10.2147/JHC.S481338
Dendritic Cell-Related Gene Signatures in Hepatocellular Carcinoma: An Analysis for Prognosis and Therapy Efficacy Evaluation
Abstract
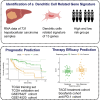
Background: This study aimed to identify dendritic cells (DCs) related genes in hepatocellular carcinoma (HCC) patients, establish DC-related subtypes and signatures, and correlate them with prognosis and treatment response.
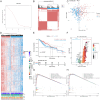
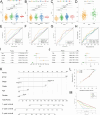
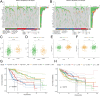
Methods: DC-related genes were screened using Weighted Gene Co-expression Network Analysis (WGCNA) based on RNA sequencing from the TCGA (374 samples), GSE14520 (242 samples), and GSE76427 datasets (115 samples), following immune infiltration assessment by the TIME method. Two DC-related subtypes in HCC were identified through unsupervised clustering. A DC-related signature (DCRS) predictive of overall survival was constructed using LASSO and Cox regression models, and validated across the three datasets. Additionally, genetic mutation characteristics, immune infiltration levels, and treatment sensitivity were explored in DCRS risk groups. The expression levels of DCRS genes and risk scores were validated in the transcriptome of 13 HCC patients receiving combined targeted therapy and immunotherapy in the Guangxi cohort using Wilcoxon test.
Results: A signature consisting of 13 genes related to DCs was constructed, and the superior prognostic consistency of the low DCRS risk group was validated across the TCGA (P=0.003), GSE76427 (P=0.005), and GSE14520 (P=0.047) datasets. Furthermore, in the 147-sample transarterial chemoembolization (TACE) treatment dataset GSE104580, the response group exhibited lower risk scores than the non-response group (P=0.01), whereas in the 140-sample Sorafenib treatment dataset GSE109211 (P=0.041) and the 17-sample anti-PD-1 treatment dataset GSE202069 (P=0.027), the risk scores were higher in the response group. We also validated the gene expression levels of DCRS and the higher risk scores in the response group of the Guangxi cohort (P=0.034).
Conclusion: A DCRS consisting of 13 genes was established in HCC, facilitating the prediction of patient prognosis and responsiveness to TACE, targeted therapy, and immunotherapy.
Keywords: dendritic cells; hepatocellular carcinoma; immunotherapy; prognosis; targeted therapy.
© 2024 Huang et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

