Identification of novel small-molecule inhibitors of SARS-CoV-2 by chemical genetics
- PMID: 39309487
- PMCID: PMC11413674
- DOI: 10.1016/j.apsb.2024.05.026
Identification of novel small-molecule inhibitors of SARS-CoV-2 by chemical genetics
Abstract
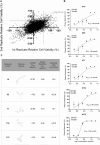
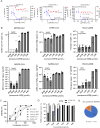
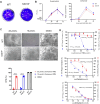
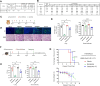
There are only eight approved small molecule antiviral drugs for treating COVID-19. Among them, four are nucleotide analogues (remdesivir, JT001, molnupiravir, and azvudine), while the other four are protease inhibitors (nirmatrelvir, ensitrelvir, leritrelvir, and simnotrelvir-ritonavir). Antiviral resistance, unfavourable drug‒drug interaction, and toxicity have been reported in previous studies. Thus there is a dearth of new treatment options for SARS-CoV-2. In this work, a three-tier cell-based screening was employed to identify novel compounds with anti-SARS-CoV-2 activity. One compound, designated 172, demonstrated broad-spectrum antiviral activity against multiple human pathogenic coronaviruses and different SARS-CoV-2 variants of concern. Mechanistic studies validated by reverse genetics showed that compound 172 inhibits the 3-chymotrypsin-like protease (3CLpro) by binding to an allosteric site and reduces 3CLpro dimerization. A drug synergistic checkerboard assay demonstrated that compound 172 can achieve drug synergy with nirmatrelvir in vitro. In vivo studies confirmed the antiviral activity of compound 172 in both Golden Syrian Hamsters and K18 humanized ACE2 mice. Overall, this study identified an alternative druggable site on the SARS-CoV-2 3CLpro, proposed a potential combination therapy with nirmatrelvir to reduce the risk of antiviral resistance and shed light on the development of allosteric protease inhibitors for treating a range of coronavirus diseases.
Keywords: 3CLpro inhibitor; Allosteric-site inhibitor; Animal models; Broad-spectrum antiviral treatment; Chemical genetics; High throughput screening; Reverse genetics; SARS-CoV-2.
© 2024 The Authors.
Conflict of interest statement
The authors have no conflict to declare.
Figures


References
-
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

