Triple three-dimensional MS/MS spectrum facilitates quantitative ginsenosides-targeted sub-metabolome characterization in notoginseng
- PMID: 39309494
- PMCID: PMC11413663
- DOI: 10.1016/j.apsb.2024.04.029
Triple three-dimensional MS/MS spectrum facilitates quantitative ginsenosides-targeted sub-metabolome characterization in notoginseng
Abstract
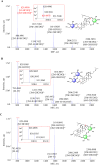
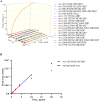
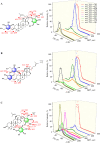
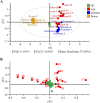
Although serving as the workhorse, MS/MS cannot fully satisfy the analytical requirements of quantitative sub-metabolome characterization. Because more information intrinsically correlates to more structural and concentration clues, here, efforts were devoted to comprehensively tracing and deciphering MS/MS behaviors through constructing triple three-dimensional (3×3D)-MS/MS spectrum. Ginsenosides-targeted metabolomics of notoginseng, one of the most famous edible medicinal plants, was employed as a proof-of-concept. Serial authentic ginsenosides were deployed to build the correlations between 3×3D-MS/MS spectra and structure/concentration features. Through assaying ginsenosides with progressive concentrations using QTOF-MS to configure 1st 3D spectrum, the generations of MS1 spectral signals, particularly multi-charged multimer anions, e.g., [2M-2H]2- and [2M+2HCOO]2- ions, relied on both concentration and the amount of sugar chains. By programming progressive collision energies to the front collision cell of Qtrap-MS device to gain 2nd 3D spectrum, optimal collision energy (OCE) corresponding to the glycosidic bond fission was primarily correlated with the masses of precursor and fragment ions and partially governed by the glycosidation site. The quantitative relationships between OCEs and masses of precursor and fragment ions were utilized to build large-scale quantitative program for ginsenosides. After applying progressive exciting energies to the back collision chamber to build 3rd 3D spectrum, the fragment ion and the decomposition product anion exhibited identical dissociation trajectories when they shared the same molecular geometry. After ginsenosides-focused quantitative metabolomics, significant differences occurred for sub-metabolome amongst different parts of notoginseng. The differential ginsenosides were confirmatively identified by applying the correlations between 3×3D-MS/MS spectra and structures. Together, 3×3D-MS/MS spectrum covers all MS/MS behaviors and dramatically facilitates sub-metabolome characterization from both quantitative program development and structural identification.
Keywords: Full exciting energy ramp-MS3 spectrum; Ginsenosides; Notoginseng; Quantitative sub-metabolome characterization; Structural identification.
© 2024 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- An N., Zhu Q.F., Wang Y.Z., Xiong C.F., Hu Y.N., Feng Y.Q. Integration of chemical derivatization and in-source fragmentation mass spectrometry for high-coverage profiling of submetabolomes. Anal Chem. 2021;93:11321–11328. - PubMed
-
- Zhao S., Li H., Han W., Chan W., Li L. Metabolomic coverage of chemical-group-submetabolome analysis: group classification and four-channel chemical isotope labeling LC‒MS. Anal Chem. 2019;91:12108–12115. - PubMed
-
- Yuan B.F., Zhu Q.F., Guo N., Zheng S.J., Wang Y.L., Wang J., et al. Comprehensive profiling of fecal metabolome of mice by integrated chemical isotope labeling-mass spectrometry analysis. Anal Chem. 2018;90:3512–3520. - PubMed
-
- Calderón-Santiago M., Priego-Capote F., Luque De Castro M.D. Enhanced detection and identification in metabolomics by use of LC–MS/MS untargeted analysis in combination with gas-phase fractionation. Anal Chem. 2014;86:7558–7565. - PubMed
-
- Chen J., Zhang P., Qin S., Tan B., Li S., Tang S., et al. Stepwise solid phase extraction integrated with chemical derivatization for all-in-one injection LC‒MS/MS analysis of metabolome and lipidome. Anal Chim Acta. 2023;1241 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

