Molecular Classification of HNSCC Based on Inflammatory Response-Related Genes - Integrated Single-Cell and Bulk RNA-Seq Analysis
- PMID: 39310901
- PMCID: PMC11414753
- DOI: 10.2147/JIR.S473823
Molecular Classification of HNSCC Based on Inflammatory Response-Related Genes - Integrated Single-Cell and Bulk RNA-Seq Analysis
Abstract
Objective: Tumor cells, inflammatory cells, and chemical factors collaboratively orchestrate a sophisticated signaling network, culminating in the formation of the inflammatory tumor microenvironment (TME). The present study sought to explore the nature of the inflammatory response in HNSCC and to decipher its influence on immunotherapeutic.
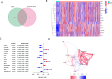
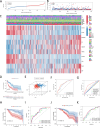
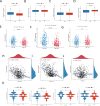
Materials and methods: A thorough analysis was performed utilizing the TCGA cohort along with two GEO cohorts. Unsupervised clustering of 200 inflammatory response-related genes (IRGs) was applied using the k-means algorithm to explore the heterogeneity of HNSCC. Additionally, a prognostic signature based on IRGs genes was constructed using Lasso regression. Meanwhiles, the expression of IRGs were identified in tumors and paracancerous tissues at the single-cell level. The crosstalk between IRGs was explored using CellChat and the patterns of incoming and outgoing signals were identified. Finally, qPCR was used to verify the expression of hub genes.
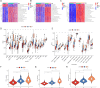
Results: There were significant differences in immune-cell function and immune-cell infiltration among three inflammatory response clusters. Additionally, we also constructed a prognostic model which could predicted the responses of common chemotherapeutic drugs and immunotherapy. Furthermore, qPCR and sc-RNA seq corroborated that the expression profiles of the prognostic genes were largely in alignment with the findings from the bioinformatics analysis. Ultimately, the molecular docking demonstrated favorable binding affinities between the pivotal gene-SCC7 and four chemotherapeutic drugs.
Conclusion: This research has uniquely shed light on the intricate connection between the inflammatory response profiles and the immune infiltration patterns in HNSCC.
Keywords: HNSCC; TME; immunotherapy; inflammatory response; sc-RNA seq.
© 2024 Zhu et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures





Similar articles
-
Identification and validation of a novel signature based on cell-cell communication in head and neck squamous cell carcinoma by integrated analysis of single-cell transcriptome and bulk RNA-sequencing.Front Oncol. 2023 May 5;13:1136729. doi: 10.3389/fonc.2023.1136729. eCollection 2023. Front Oncol. 2023. PMID: 37213285 Free PMC article.
-
Development of an Immunogenomic Landscape-Based Prognostic Index of Head and Neck Squamous Cell Carcinoma.Front Mol Biosci. 2020 Nov 24;7:586344. doi: 10.3389/fmolb.2020.586344. eCollection 2020. Front Mol Biosci. 2020. PMID: 33330624 Free PMC article.
-
Development and validation a prognostic model based on natural killer T cells marker genes for predicting prognosis and characterizing immune status in glioblastoma through integrated analysis of single-cell and bulk RNA sequencing.Funct Integr Genomics. 2023 Aug 31;23(3):286. doi: 10.1007/s10142-023-01217-7. Funct Integr Genomics. 2023. PMID: 37650991
-
Establishment of an ovarian cancer omentum metastasis-related prognostic model by integrated analysis of scRNA-seq and bulk RNA-seq.J Ovarian Res. 2022 Nov 23;15(1):123. doi: 10.1186/s13048-022-01059-0. J Ovarian Res. 2022. PMID: 36424614 Free PMC article.
-
The Landscape of the Tumor Microenvironment in Skin Cutaneous Melanoma Reveals a Prognostic and Immunotherapeutically Relevant Gene Signature.Front Cell Dev Biol. 2021 Oct 1;9:739594. doi: 10.3389/fcell.2021.739594. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34660598 Free PMC article.
References
LinkOut - more resources
Full Text Sources

