Circulating miRNAs in the Plasma of Post-COVID-19 Patients with Typical Recovery and Those with Long-COVID Symptoms: Regulation of Immune Response-Associated Pathways
- PMID: 39311385
- PMCID: PMC11417918
- DOI: 10.3390/ncrna10050048
Circulating miRNAs in the Plasma of Post-COVID-19 Patients with Typical Recovery and Those with Long-COVID Symptoms: Regulation of Immune Response-Associated Pathways
Abstract
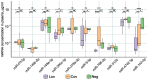
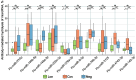
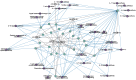
Following the acute phase of SARS-CoV-2 infection, certain individuals experience persistent symptoms referred to as long COVID. This study analyzed the patients categorized into three distinct groups: (1) individuals presenting rheumatological symptoms associated with long COVID, (2) patients who have successfully recovered from COVID-19, and (3) donors who have never contracted COVID-19. A notable decline in the expression of miR-200c-3p, miR-766-3p, and miR-142-3p was identified among patients exhibiting rheumatological symptoms of long COVID. The highest concentration of miR-142-3p was found in healthy donors. One potential way to reduce miRNA concentrations is through antibody-mediated hydrolysis. Not only can antibodies possessing RNA-hydrolyzing activity recognize the miRNA substrate specifically, but they also catalyze its hydrolysis. The analysis of the catalytic activity of plasma antibodies revealed that antibodies from patients with long COVID demonstrated lower hydrolysis activity against five fluorescently labeled oligonucleotide sequences corresponding to the Flu-miR-146b-5p, Flu-miR-766-3p, Flu-miR-4742-3p, and Flu-miR-142-3p miRNAs and increased activity against the Flu-miR-378a-3p miRNA compared to other patient groups. The changes in miRNA concentrations and antibody-mediated hydrolysis of miRNAs are assumed to have a complex regulatory mechanism that is linked to gene pathways associated with the immune system. We demonstrate that all six miRNAs under analysis are associated with a large number of signaling pathways associated with immune response-associated pathways.
Keywords: COVID-19; RNA hydrolysis; SARS-CoV-2; antibodies; catalytic antibodies; long COVID; miR-142-3p; miR-200c-3p; miR-766-3p; miRNA; pathways; regulatory network; rheumatologically.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Differential microRNA expression in the peripheral blood from human patients with COVID-19.J Clin Lab Anal. 2020 Oct;34(10):e23590. doi: 10.1002/jcla.23590. Epub 2020 Sep 22. J Clin Lab Anal. 2020. PMID: 32960473 Free PMC article.
-
SIV Infection Regulates Compartmentalization of Circulating Blood Plasma miRNAs within Extracellular Vesicles (EVs) and Extracellular Condensates (ECs) and Decreases EV-Associated miRNA-128.Viruses. 2023 Feb 24;15(3):622. doi: 10.3390/v15030622. Viruses. 2023. PMID: 36992331 Free PMC article.
-
Evaluation of miR-200c-3p and miR-421-5p levels during immune responses in the admitted and recovered COVID-19 subjects.Infect Genet Evol. 2022 Mar;98:105207. doi: 10.1016/j.meegid.2022.105207. Epub 2022 Jan 5. Infect Genet Evol. 2022. PMID: 34999004 Free PMC article.
-
Differentially expressed plasmatic microRNAs in Brazilian patients with Coronavirus disease 2019 (COVID-19): preliminary results.Mol Biol Rep. 2022 Jul;49(7):6931-6943. doi: 10.1007/s11033-022-07338-9. Epub 2022 Mar 17. Mol Biol Rep. 2022. PMID: 35301654 Free PMC article.
-
Breast milk microRNAs: Potential players in oral tolerance development.Front Immunol. 2023 Mar 14;14:1154211. doi: 10.3389/fimmu.2023.1154211. eCollection 2023. Front Immunol. 2023. PMID: 36999032 Free PMC article. Review.
Cited by
-
Vaxtherapy, a Multiphase Therapeutic Protocol Approach for Longvax, the COVID-19 Vaccine-Induced Disease: Spike Persistence as the Core Culprit and Its Downstream Effects.Diseases. 2025 Jun 30;13(7):204. doi: 10.3390/diseases13070204. Diseases. 2025. PMID: 40709992 Free PMC article.
-
MicroRNAs in long COVID: roles, diagnostic biomarker potential and detection.Hum Genomics. 2025 Aug 13;19(1):90. doi: 10.1186/s40246-025-00810-0. Hum Genomics. 2025. PMID: 40804645 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

