Atomistic simulations reveal impacts of missense mutations on the structure and function of SynGAP1
- PMID: 39311700
- PMCID: PMC11418247
- DOI: 10.1093/bib/bbae458
Atomistic simulations reveal impacts of missense mutations on the structure and function of SynGAP1
Abstract
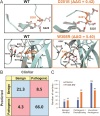
De novo mutations in the synaptic GTPase activating protein (SynGAP) are associated with neurological disorders like intellectual disability, epilepsy, and autism. SynGAP is also implicated in Alzheimer's disease and cancer. Although pathogenic variants are highly penetrant in neurodevelopmental conditions, a substantial number of them are caused by missense mutations that are difficult to diagnose. Hence, in silico mutagenesis was performed for probing the missense effects within the N-terminal region of SynGAP structure. Through extensive molecular dynamics simulations, encompassing three 150-ns replicates for 211 variants, the impact of missense mutations on the protein fold was assessed. The effect of the mutations on the folding stability was also quantitatively assessed using free energy calculations. The mutations were categorized as potentially pathogenic or benign based on their structural impacts. Finally, the study introduces wild-type-SynGAP in complex with RasGTPase at the inner membrane, while considering the potential effects of mutations on these key interactions. This study provides structural perspective to the clinical assessment of SynGAP missense variants and lays the foundation for future structure-based drug discovery.
Keywords: SynGAP1; in silico mutagenesis; intellectual disability; missense mutation; molecular dynamics (MD) simulation; structural bioinformatics.
© The Author(s) 2024. Published by Oxford University Press.
Figures


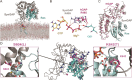

Similar articles
-
Multi-parametric analysis of 57 SYNGAP1 variants reveal impacts on GTPase signaling, localization, and protein stability.Am J Hum Genet. 2021 Jan 7;108(1):148-162. doi: 10.1016/j.ajhg.2020.11.011. Epub 2020 Dec 11. Am J Hum Genet. 2021. PMID: 33308442 Free PMC article.
-
Key roles of C2/GAP domains in SYNGAP1-related pathophysiology.Cell Rep. 2024 Sep 24;43(9):114733. doi: 10.1016/j.celrep.2024.114733. Epub 2024 Sep 12. Cell Rep. 2024. PMID: 39269903
-
Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency.Hum Mutat. 2013 Feb;34(2):385-94. doi: 10.1002/humu.22248. Epub 2012 Dec 12. Hum Mutat. 2013. PMID: 23161826
-
Species-conserved SYNGAP1 phenotypes associated with neurodevelopmental disorders.Mol Cell Neurosci. 2018 Sep;91:140-150. doi: 10.1016/j.mcn.2018.03.008. Epub 2018 Mar 24. Mol Cell Neurosci. 2018. PMID: 29580901 Free PMC article. Review.
-
SYNGAP1 Syndrome and the Brain Gene Registry.Genes (Basel). 2025 Mar 30;16(4):405. doi: 10.3390/genes16040405. Genes (Basel). 2025. PMID: 40282364 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

