Characterization of a ferroptosis-related gene signature predicting survival and immunotherapeutic response in lung adenocarcinoma
- PMID: 39311766
- PMCID: PMC11466487
- DOI: 10.18632/aging.206110
Characterization of a ferroptosis-related gene signature predicting survival and immunotherapeutic response in lung adenocarcinoma
Abstract
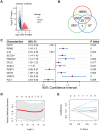
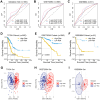
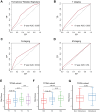
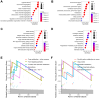
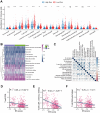
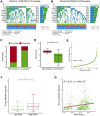
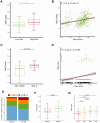
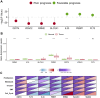
Lung cancer remains the leading cause of cancer-related death worldwide, and drug resistance represents the main obstacle responsible for the poor mortality and prognosis. Here, to identify a novel gene signature for predicting survival and drug response, we jointly investigated RNA sequencing data of lung adenocarcinoma patients from TCGA and GEO databases, and identified a ferroptosis-related gene signature. The signature was validated in the validation set and two external cohorts. The high-risk group had a reduced survival than the low-risk group (P < 0.05). Moreover, the established gene signature was associated with tumor mutation burden, microsatellite instability, and response to immune checkpoint blockade. In addition, four candidate oncogenes (RRM2, SLC2A1, DDIT4, and VDAC2) were identified to be candidate oncogenes using in silico and wet experiments, which could serve as potential therapeutic targets. Collectively, this study developed a novel ferroptosis-related gene signature for predicting prognosis and drug response, and identified four candidate oncogenes for lung adenocarcinoma.
Keywords: drug response; ferroptosis; gene signature; lung adenocarcinoma; prognosis.
Conflict of interest statement
Figures

References
-
- Machtay M, Duan F, Siegel BA, Snyder BS, Gorelick JJ, Reddin JS, Munden R, Johnson DW, Wilf LH, DeNittis A, Sherwin N, Cho KH, Kim SK, et al.. Prediction of survival by [18F]fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: results of the ACRIN 6668/RTOG 0235 trial. J Clin Oncol. 2013; 31:3823–30. 10.1200/JCO.2012.47.5947 - DOI - PMC - PubMed
-
- Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B, Wu DS, Fang ZY, Bao Y, Jiang MM, Yuan JH, Qu L, Wang LH. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017; 16:111. 10.1186/s12943-017-0681-0 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

