Use of the Novel Site-Directed Enzyme Enhancement Therapy (SEE-Tx) Drug Discovery Platform to Identify Pharmacological Chaperones for Glutaric Acidemia Type 1
- PMID: 39312412
- PMCID: PMC11472340
- DOI: 10.1021/acs.jmedchem.4c00292
Use of the Novel Site-Directed Enzyme Enhancement Therapy (SEE-Tx) Drug Discovery Platform to Identify Pharmacological Chaperones for Glutaric Acidemia Type 1
Abstract
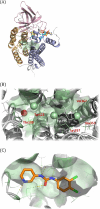
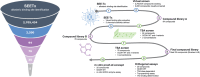
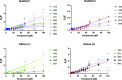
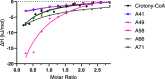
Allosteric regulators acting as pharmacological chaperones hold promise for innovative therapeutics since they target noncatalytic sites and stabilize the folded protein without competing with the natural substrate, resulting in a net gain of function. Exogenous allosteric regulators are typically more selective than active site inhibitors and can be more potent than competitive inhibitors when the natural substrate levels are high. To identify novel structure-targeted allosteric regulators (STARs) that bind to and stabilize the mitochondrial enzyme glutaryl-CoA dehydrogenase (GCDH), the computational site-directed enzyme enhancement therapy (SEE-Tx) technology was applied. SEE-Tx is an innovative drug discovery platform with the potential to identify drugs for treating protein misfolding disorders, such as glutaric acidemia type 1 (GA1) disease. Putative allosteric regulators were discovered using structure- and ligand-based virtual screening methods and validated using orthogonal biophysical and biochemical assays. The computational approach presented here could be used to discover allosteric regulators of other protein misfolding disorders.
Conflict of interest statement
The authors declare the following competing financial interest(s): EC, AD, AMG, MM, and XB completed the research and the authorship of this article within the scope of their employ-ment. EC, AD, AMG, and XB are employees of Gain Therapeu-tics Sucursal en Espaa and MM is an employee and sharehold-er of Minoryx Therapeutics. ACM and SWG are co-founders and shareholders of iniuva GmbH.
Figures

References
-
- Kölker S.; Garbade S. F.; Greenberg C. R.; Leonard J. V.; Saudubray J. M.; Ribes A.; Kalkanoglu H. S.; Lund A. M.; Merinero B.; Wajner M.; et al. Natural History, Outcome, and Treatment Efficacy in Children and Adults with Glutaryl-CoA Dehydrogenase Deficiency. Pediatr. Res. 2006, 59 (6), 840–847. 10.1203/01.pdr.0000219387.79887.86. - DOI - PubMed
-
- Boy N.; Mühlhausen C.; Maier E. M.; Heringer J.; Assmann B.; Burgard P.; Dixon M.; Fleissner S.; Greenberg C. R.; Harting I.; et al. Proposed Recommendations for Diagnosing and Managing Individuals with Glutaric Aciduria Type I: Second Revision. J. Inherit. Metab. Dis. 2017, 40 (1), 75–101. 10.1007/s10545-016-9999-9. - DOI - PubMed
-
- Strauss K. A.; Williams K. B.; Carson V. J.; Poskitt L.; Bowser L. E.; Young M.; Robinson D. L.; Hendrickson C.; Beiler K.; Taylor C. M.; et al. Glutaric Acidemia Type 1: Treatment and Outcome of 168 Patients over Three Decades. Mol. Genet. Metab. 2020, 131 (3), 325–340. 10.1016/j.ymgme.2020.09.007. - DOI - PubMed
-
- Märtner E. M. C.; Thimm E.; Guder P.; Schiergens K. A.; Rutsch F.; Roloff S.; Marquardt I.; Das A. M.; Freisinger P.; Grünert S. C.; et al. The Biochemical Subtype is a Predictor for Cognitive Function in Glutaric Aciduria Type 1: A National Prospective Follow-Up Study. Sci. Rep. 2021, 11 (1), 1930010.1038/s41598-021-98809-9. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

