Multiple lines of evidence for disruption of nuclear lamina and nucleoporins in FUS amyotrophic lateral sclerosis
- PMID: 39312484
- PMCID: PMC11684083
- DOI: 10.1093/brain/awae224
Multiple lines of evidence for disruption of nuclear lamina and nucleoporins in FUS amyotrophic lateral sclerosis
Abstract
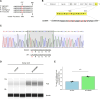
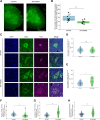
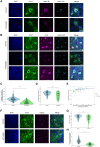
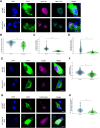
Advanced pathological and genetic approaches have revealed that mutations in fused in sarcoma/translated in liposarcoma (FUS/TLS), which is pivotal for DNA repair, alternative splicing, translation and RNA transport, cause familial amyotrophic lateral sclerosis (ALS). The generation of suitable animal models for ALS is essential for understanding its pathogenesis and developing therapies. Therefore, we used CRISPR-Cas9 to generate FUS-ALS mutation in the non-classical nuclear localization signal (NLS), H517D (mouse position: H509D) and genome-edited mice. Fus WT/H509D mice showed progressive motor impairment (accelerating rotarod and DigiGait system) with age, which was associated with the loss of motor neurons and disruption of the nuclear lamina and nucleoporins and DNA damage in spinal cord motor neurons. We confirmed the validity of our model by showing that nuclear lamina and nucleoporin disruption were observed in lower motor neurons differentiated from patient-derived human induced pluripotent stem cells (hiPSC-LMNs) with FUS-H517D and in the post-mortem spinal cord of patients with ALS. RNA sequence analysis revealed that most nuclear lamina and nucleoporin-linking genes were significantly decreased in FUS-H517D hiPSC-LMNs. This evidence suggests that disruption of the nuclear lamina and nucleoporins is crucial for ALS pathomechanisms. Combined with patient-derived hiPSC-LMNs and autopsy samples, this mouse model might provide a more reliable understanding of ALS pathogenesis and might aid in the development of therapeutic strategies.
Keywords: FUS; amyotrophic lateral sclerosis; nuclear lamina; nuclear pore complex.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
H.O. reports grants and personal fees as a Chief Scientific Officer from K Pharma, Inc. during the conduct of the study; personal fees from Sanbio Co. Ltd., outside the submitted work. In addition, H.O. has a patent on a therapeutic agent for amyotrophic lateral sclerosis and composition for treatment licensed to K Pharma, Inc. The other authors have declared that no conflict of interest exists.
Figures



References
-
- Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. - PubMed
-
- Zou Z-Y, Liu M-S, Li X-G, Cui L-Y. Mutations in FUS are the most frequent genetic cause in juvenile sporadic ALS patients of Chinese origin. Amyotroph Lateral Scler Frontotemporal Degener. 2016;17(3–4):249–252. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

