Deep Profiling of Plasma Proteoforms with Engineered Nanoparticles for Top-Down Proteomics
- PMID: 39312774
- PMCID: PMC11789057
- DOI: 10.1021/acs.jproteome.4c00621
Deep Profiling of Plasma Proteoforms with Engineered Nanoparticles for Top-Down Proteomics
Abstract
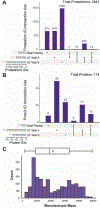
The dynamic range challenge for the detection of proteins and their proteoforms in human plasma has been well documented. Here, we use the nanoparticle protein corona approach to enrich low-abundance proteins selectively and reproducibly from human plasma and use top-down proteomics to quantify differential enrichment for the 2841 detected proteoforms from 114 proteins. Furthermore, nanoparticle enrichment allowed top-down detection of proteoforms between ∼1 μg/mL and ∼10 pg/mL in absolute abundance, providing up to a 105-fold increase in proteome depth over neat plasma in which only proteoforms from abundant proteins (>1 μg/mL) were detected. The ability to monitor medium and some low-abundant proteoforms through reproducible enrichment significantly extends the applicability of proteoform research by adding depth beyond albumin, immunoglobins, and apolipoproteins to uncover many involved in immunity and cell signaling. As proteoforms carry unique information content relative to peptides, this report opens the door to deeper proteoform sequencing in clinical proteomics of disease or aging cohorts.
Keywords: nanoparticles; plasma; protein corona; proteoforms; top-down proteomics.
Figures



Update of
-
Deep Profiling of Plasma Proteoforms with Engineered Nanoparticles for Top-down Proteomics.bioRxiv [Preprint]. 2024 Jul 21:2024.07.20.604425. doi: 10.1101/2024.07.20.604425. bioRxiv. 2024. Update in: J Proteome Res. 2024 Oct 4;23(10):4694-4703. doi: 10.1021/acs.jproteome.4c00621. PMID: 39071411 Free PMC article. Updated. Preprint.
References
-
- Surinova S; Schiess R; Huttenhain R; Cerciello F; Wollscheid B; Aebersold R On the development of plasma protein biomarkers. J. Proteome Res 2011, 10 (1), 5–16. - PubMed
-
- Hortin GL; Sviridov D The dynamic range problem in the analysis of the plasma proteome. J. Proteomics 2010, 73 (3), 629–636. - PubMed
-
- Tirumalai RS; Chan KC; Prieto DA; Issaq HJ; Conrads TP; Veenstra TD Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics 2003, 2 (10), 1096–1103. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

