Spliceosomal vulnerability of MYCN-amplified neuroblastoma is contingent on PRMT5-mediated regulation of epitranscriptomic and metabolomic pathways
- PMID: 39313128
- PMCID: PMC7618037
- DOI: 10.1016/j.canlet.2024.217263
Spliceosomal vulnerability of MYCN-amplified neuroblastoma is contingent on PRMT5-mediated regulation of epitranscriptomic and metabolomic pathways
Abstract
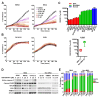
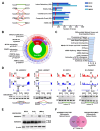
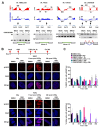
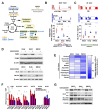
Approximately 50 % of poor prognosis neuroblastomas arise due to MYCN over-expression. We previously demonstrated that MYCN and PRMT5 proteins interact and PRMT5 knockdown led to apoptosis of MYCN-amplified (MNA) neuroblastoma. Here we evaluate the highly selective first-in-class PRMT5 inhibitor GSK3203591 and its in vivo analogue GSK3326593 as targeted therapeutics for MNA neuroblastoma. Cell-line analyses show MYCN-dependent growth inhibition and apoptosis, with approximately 200-fold greater sensitivity of MNA neuroblastoma lines. RNA sequencing of three MNA neuroblastoma lines treated with GSK3203591 reveal deregulated MYCN transcriptional programmes and altered mRNA splicing, converging on key regulatory pathways such as DNA damage response, epitranscriptomics and cellular metabolism. Stable isotope labelling experiments in the same cell lines demonstrate that glutamine metabolism is impeded following GSK3203591 treatment, linking with disruption of the MLX/Mondo nutrient sensors via intron retention of MLX mRNA. Interestingly, glutaminase (GLS) protein decreases after GSK3203591 treatment despite unchanged transcript levels. We demonstrate that the RNA methyltransferase METTL3 and cognate reader YTHDF3 proteins are lowered following their mRNAs undergoing GSK3203591-induced splicing alterations, indicating epitranscriptomic regulation of GLS; accordingly, we observe decreases of GLS mRNA m6A methylation following GSK3203591 treatment, and decreased GLS protein following YTHDF3 knockdown. In vivo efficacy of GSK3326593 is confirmed by increased survival of Th-MYCN mice, with drug treatment triggering splicing events and protein decreases consistent with in vitro data. Together our study demonstrates the PRMT5-dependent spliceosomal vulnerability of MNA neuroblastoma and identifies the epitranscriptome and glutamine metabolism as critical determinants of this sensitivity.
Crown Copyright © 2024. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. - PubMed
-
- Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47(12):1411–1414. - PubMed
-
- Zimmerman MW, Liu Y, He S, Durbin AD, Abraham BJ, Easton J, et al. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 2018;8(3):320–335. doi: 10.1158/2159-8290.CD-17-0993. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

