Large B-cell lymphomas with CCND1 rearrangement have different immunoglobulin gene breakpoints and genomic profile than mantle cell lymphoma
- PMID: 39313500
- PMCID: PMC11420347
- DOI: 10.1038/s41408-024-01146-z
Large B-cell lymphomas with CCND1 rearrangement have different immunoglobulin gene breakpoints and genomic profile than mantle cell lymphoma
Abstract
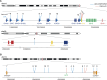
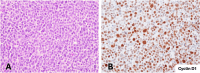
Mantle cell lymphoma (MCL) is genetically characterized by the IG::CCND1 translocation mediated by an aberrant V(D)J rearrangement. CCND1 translocations and overexpression have been identified in occasional aggressive B-cell lymphomas with unusual features for MCL. The mechanism generating CCND1 rearrangements in these tumors and their genomic profile are not known. We have reconstructed the IG::CCND1 translocations and the genomic profile of 13 SOX11-negative aggressive B-cell lymphomas using whole genome/exome and target sequencing. The mechanism behind the translocation was an aberrant V(D)J rearrangement in three tumors and by an anomalous IGH class-switch recombination (CSR) or somatic hypermutation (SHM) mechanism in ten. The tumors with a V(D)J-mediated translocation were two blastoid MCL and one high-grade B-cell lymphoma. None of them had a mutational profile suggestive of DLBCL. The ten tumors with CSR/SHM-mediated IGH::CCND1 were mainly large B-cell lymphomas, with mutated genes commonly seen in DLBCL and BCL6 rearrangements in 6. Two cases, which transformed from marginal zone lymphomas, carried mutations in KLF2, TNFAIP3 and KMT2D. These findings expand the spectrum of tumors carrying CCND1 rearrangement that may occur as a secondary event in DLBCL mediated by aberrant CSR/SHM and associated with a mutational profile different from that of MCL.
© 2024. The Author(s).
Conflict of interest statement
F.N. received honoraria from Janssen, AbbVie, AstraZeneca, and SOPHiA GENETICS for speaking in educational activities, and research funding from Gilead. E.C. has been a consultant for Takeda; has received honoraria from Janssen, EUSA Pharma and Roche for speaking at educational activities and research funding from AstraZeneca and is an inventor on 2 patents filed by the National Institutes of Health, National Cancer Institute: “Methods for selecting and treating lymphoma types,” licensed to NanoString Technologies, and “Evaluation of MCL and methods related thereof”, not related to this project. F.N. and E.C. licensed the use of the protected IgCaller algorithm for Diagnóstica Longwood. The remaining authors declare no competing financial interests.
Figures



References
-
- Ok CY, Xu-Monette ZY, Tzankov A, O’Malley DP, Montes-Moreno S, Visco C, et al. Prevalence and clinical implications of cyclin D1 expression in diffuse large B-cell lymphoma (DLBCL) treated with immunochemotherapy: a report from the International DLBCL Rituximab-CHOP Consortium Program. Cancer. 2014;120:1818–29. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

