Beyond the usual suspects: multi-factorial computational models in the search for neurodegenerative disease mechanisms
- PMID: 39313512
- PMCID: PMC11420368
- DOI: 10.1038/s41398-024-03073-w
Beyond the usual suspects: multi-factorial computational models in the search for neurodegenerative disease mechanisms
Abstract
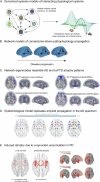
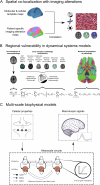
From Alzheimer's disease to amyotrophic lateral sclerosis, the molecular cascades underlying neurodegenerative disorders remain poorly understood. The clinical view of neurodegeneration is confounded by symptomatic heterogeneity and mixed pathology in almost every patient. While the underlying physiological alterations originate, proliferate, and propagate potentially decades before symptomatic onset, the complexity and inaccessibility of the living brain limit direct observation over a patient's lifespan. Consequently, there is a critical need for robust computational methods to support the search for causal mechanisms of neurodegeneration by distinguishing pathogenic processes from consequential alterations, and inter-individual variability from intra-individual progression. Recently, promising advances have been made by data-driven spatiotemporal modeling of the brain, based on in vivo neuroimaging and biospecimen markers. These methods include disease progression models comparing the temporal evolution of various biomarkers, causal models linking interacting biological processes, network propagation models reproducing the spatial spreading of pathology, and biophysical models spanning cellular- to network-scale phenomena. In this review, we discuss various computational approaches for integrating cross-sectional, longitudinal, and multi-modal data, primarily from large observational neuroimaging studies, to understand (i) the temporal ordering of physiological alterations, i(i) their spatial relationships to the brain's molecular and cellular architecture, (iii) mechanistic interactions between biological processes, and (iv) the macroscopic effects of microscopic factors. We consider the extents to which computational models can evaluate mechanistic hypotheses, explore applications such as improving treatment selection, and discuss how model-informed insights can lay the groundwork for a pathobiological redefinition of neurodegenerative disorders.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Imaging plus X: multimodal models of neurodegenerative disease.Curr Opin Neurol. 2017 Aug;30(4):371-379. doi: 10.1097/WCO.0000000000000460. Curr Opin Neurol. 2017. PMID: 28520598 Free PMC article. Review.
-
Multifactorial causal model of brain (dis)organization and therapeutic intervention: Application to Alzheimer's disease.Neuroimage. 2017 May 15;152:60-77. doi: 10.1016/j.neuroimage.2017.02.058. Epub 2017 Feb 28. Neuroimage. 2017. PMID: 28257929
-
Computational Causal Modeling of the Dynamic Biomarker Cascade in Alzheimer's Disease.Comput Math Methods Med. 2019 Feb 3;2019:6216530. doi: 10.1155/2019/6216530. eCollection 2019. Comput Math Methods Med. 2019. PMID: 30863455 Free PMC article.
-
Personalized brain models identify neurotransmitter receptor changes in Alzheimer's disease.Brain. 2022 Jun 3;145(5):1785-1804. doi: 10.1093/brain/awab375. Brain. 2022. PMID: 34605898 Free PMC article.
-
Understanding disease progression and improving Alzheimer's disease clinical trials: Recent highlights from the Alzheimer's Disease Neuroimaging Initiative.Alzheimers Dement. 2019 Jan;15(1):106-152. doi: 10.1016/j.jalz.2018.08.005. Epub 2018 Oct 13. Alzheimers Dement. 2019. PMID: 30321505 Review.
Cited by
-
Artificial Intelligence and Neuroscience: Transformative Synergies in Brain Research and Clinical Applications.J Clin Med. 2025 Jan 16;14(2):550. doi: 10.3390/jcm14020550. J Clin Med. 2025. PMID: 39860555 Free PMC article. Review.
-
Towards advanced regenerative therapeutics to tackle cardio-cerebrovascular diseases.Am Heart J Plus. 2025 Mar 1;53:100520. doi: 10.1016/j.ahjo.2025.100520. eCollection 2025 May. Am Heart J Plus. 2025. PMID: 40230658 Free PMC article. Review.
-
Glucose metabolism alterations and Aβ deposition in AD and FTD are related to the distribution of neurotransmitter systems.Eur J Nucl Med Mol Imaging. 2025 Aug 7. doi: 10.1007/s00259-025-07485-8. Online ahead of print. Eur J Nucl Med Mol Imaging. 2025. PMID: 40773016
-
Implementing a tridimensional diagnostic framework for personalized medicine in neurodegenerative diseases.Alzheimers Dement. 2025 Feb;21(2):e14591. doi: 10.1002/alz.14591. Alzheimers Dement. 2025. PMID: 39976261 Free PMC article.
-
Brain-Inspired Multisensory Learning: A Systematic Review of Neuroplasticity and Cognitive Outcomes in Adult Multicultural and Second Language Acquisition.Biomimetics (Basel). 2025 Jun 12;10(6):397. doi: 10.3390/biomimetics10060397. Biomimetics (Basel). 2025. PMID: 40558367 Free PMC article. Review.
References
-
- Olanow CW, Kieburtz K, Schapira AH. Why have we failed to achieve neuroprotection in Parkinson’s disease? Ann Neurol: Off J Am Neurol Assoc Child Neurol Soc. 2008;64:S101–110. - PubMed
-
- Van Dyck CH, Swanson CJ, Aisen P, Bateman RJ, Chen C, Gee M, et al. Lecanemab in early Alzheimer’s disease. N. Engl J Med. 2023;388:9–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

