Molecular profiling of BRAF-V600E-mutant metastatic colorectal cancer in the phase 3 BEACON CRC trial
- PMID: 39313594
- PMCID: PMC11564101
- DOI: 10.1038/s41591-024-03235-9
Molecular profiling of BRAF-V600E-mutant metastatic colorectal cancer in the phase 3 BEACON CRC trial
Abstract
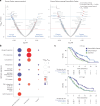
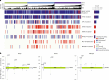
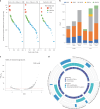
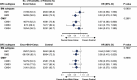
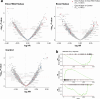
The BEACON CRC study demonstrated that encorafenib (Enco)+cetuximab (Cetux)±binimetinib (Bini) significantly improved overall survival (OS) versus Cetux + chemotherapy in previously treated patients with BRAF-V600E-mutant mCRC, providing the basis for the approval of the Enco+Cetux regimen in the United States and the European Union. A greater understanding of biomarkers predictive of response to Enco+Cetux±Bini treatment is of clinical relevance. In this prespecified, exploratory biomarker analysis of the BEACON CRC study, we characterize genomic and transcriptomic correlates of clinical outcomes and acquired resistance mechanisms through integrated clinical and molecular analysis, including whole-exome and -transcriptome tissue sequencing and circulating tumor DNA genomic profiling. Tumors with higher immune signatures showed a trend towards increased OS benefit with Enco+Bini+Cetux. RAS, MAP2K1 and MET alterations were most commonly acquired with Enco+Cetux±Bini, and more frequent in patients with a high baseline cell-cycle gene signature; baseline TP53 mutation was associated with acquired MET amplification. Acquired mutations were subclonal and polyclonal, with evidence of increased tumor mutation rate with Enco+Cetux±Bini and mutational signatures (SBS17a/b). These findings support treatment with Enco+Cetux±Bini for patients with BRAF-V600E-mutant mCRC and provide insights into the biology of response and resistance to MAPK-pathway-targeted therapy. ClinicalTrials.gov registration: NCT02928224.
© 2024. The Author(s).
Conflict of interest statement
Figures










References
-
- De Roock, W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol.11, 753–762 (2010). - PubMed
-
- Tabernero, J., Ros, J. & Élez, E. The evolving treatment landscape in BRAF-V600E-mutated metastatic colorectal cancer. Am. Soc. Clin. Oncol. Educ. Book42, 1–10 (2022). - PubMed
-
- Kopetz, S. et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med.381, 1632–1643 (2019). - PubMed
-
- Barras, D. et al. BRAF V600E mutant colorectal cancer subtypes based on gene expression. Clin. Cancer Res.23, 104–115 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

