Self-sustaining long-term 3D epithelioid cultures reveal drivers of clonal expansion in esophageal epithelium
- PMID: 39313617
- PMCID: PMC11525200
- DOI: 10.1038/s41588-024-01875-8
Self-sustaining long-term 3D epithelioid cultures reveal drivers of clonal expansion in esophageal epithelium
Abstract
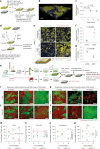
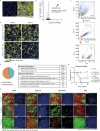
Aging epithelia are colonized by somatic mutations, which are subjected to selection influenced by intrinsic and extrinsic factors. The lack of suitable culture systems has slowed the study of this and other long-term biological processes. Here, we describe epithelioids, a facile, cost-effective method of culturing multiple mouse and human epithelia. Esophageal epithelioids self-maintain without passaging for at least 1 year, maintaining a three-dimensional structure with proliferative basal cells that differentiate into suprabasal cells, which eventually shed and retain genomic stability. Live imaging over 5 months showed that epithelioids replicate in vivo cell dynamics. Epithelioids support genetic manipulation and enable the study of mutant cell competition and selection in three-dimensional epithelia, and show how anti-cancer treatments modulate competition between transformed and wild-type cells. Finally, a targeted CRISPR-Cas9 screen shows that epithelioids recapitulate mutant gene selection in aging human esophagus and identifies additional drivers of clonal expansion, resolving the genetic networks underpinning competitive fitness.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
-
- Marchetti, M., Caliot, E. & Pringault, E. Chronic acid exposure leads to activation of the cdx2 intestinal homeobox gene in a long-term culture of mouse esophageal keratinocytes. J. Cell Sci.116, 1429–1436 (2003). - PubMed
-
- Compton, C. C., Warland, G., Nakagawa, H., Opitz, O. G. & Rustgi, A. K. Cellular characterization and successful transfection of serially subcultured normal human esophageal keratinocytes. J. Cell. Physiol.177, 274–281 (1998). - PubMed
-
- Doran, T. I., Vidrich, A. & Sun, T.-T. Intrinsic and extrinsic regulation of the differentiation of skin, corneal and esophageal epithelial cells. Cell22, 17–25 (1980). - PubMed
MeSH terms
Grants and funding
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
- 23-0063/AICR_/Worldwide Cancer Research/United Kingdom
- NC/X000885/1/National Centre for the Replacement Refinement and Reduction of Animals in Research (NC3Rs)
- C609/A17257/Cancer Research UK (CRUK)
- 105942/Z/14/A/Royal Society
- RPG-2023-136/Leverhulme Trust
- EMBO ALTF885-2015/European Molecular Biology Organization (EMBO)
- 296194/Wellcome Trust (Wellcome)
- MR/P019013/1/RCUK | Medical Research Council (MRC)
- PID2020-116163GA-I00/Agencia Canaria de Investigación, Innovación y Sociedad de la Información (ACIISI)
- 108413/A/15/D/Wellcome Trust (Wellcome)
- 105942/Z/14/Z/Royal Society
- C17918/A28870/Cancer Research UK (CRUK)
- WT_/Wellcome Trust/United Kingdom
- C609/A27326/Cancer Research UK (CRUK)
- 323267/EC | EC Seventh Framework Programm | FP7 Nuclear Fission, Safety and Radiation Protection (FP7-EURATOM-FISSION - EURATOM: Nuclear Fission and Radiation Protection)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

