Cavity-Based Discovery of New Fatty Acid Photodecarboxylases
- PMID: 39314172
- PMCID: PMC11664916
- DOI: 10.1002/cbic.202400631
Cavity-Based Discovery of New Fatty Acid Photodecarboxylases
Abstract
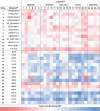
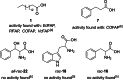
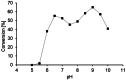
Light-dependent fatty acid photodecarboxylases (FAPs) hold significant potential for biotechnology, due to their capability to produce alka(e)nes directly from the corresponding (un)saturated natural fatty acids requiring light as the only reagent. This study expands the family of FAPs through cavity-based enzyme discovery methods. Thirty enzyme candidates with potential photodecarboxylation activity were identified by matching the cavities of four related template structures against the Protein Data Bank's flavoproteins, a library of proteins identified via the Foldseek Search Server, and homology models of sequences resulting from BLAST. Subsequent docking experiments narrowed this library to ten promising enzymes, which were expressed and assessed in vitro, identifying four photodecarboxylases. Out of these enzymes, the GMC oxidoreductase from Coccomyxa sp. Obi (CoFAP) was characterized in detail, which revealed high activity in the decarboxylation reactions of palmitic acid and octanoic acid and a broad pH tolerance (pH 6.5-9.5).
Keywords: Biocatalysis; Enzyme discovery; Photocatalysis; Photodecarboxylase.
© 2024 The Author(s). ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Radical-based photoinactivation of fatty acid photodecarboxylases.Anal Biochem. 2020 Jul 1;600:113749. doi: 10.1016/j.ab.2020.113749. Epub 2020 Apr 26. Anal Biochem. 2020. PMID: 32348726
-
Stabilisation of the Fatty Acid Decarboxylase from Chlorella variabilis by Caprylic Acid.Chembiochem. 2021 Jul 15;22(14):2420-2423. doi: 10.1002/cbic.202100182. Epub 2021 Jun 1. Chembiochem. 2021. PMID: 34002919 Free PMC article.
-
Rates of fatty acid oxidations by P450 compound I are pH dependent.Chembiochem. 2012 Sep 24;13(14):2061-4. doi: 10.1002/cbic.201200387. Epub 2012 Aug 13. Chembiochem. 2012. PMID: 22890798 Free PMC article.
-
Photoenzymes and Related Topics: An Update.Photochem Photobiol. 2018 May;94(3):459-465. doi: 10.1111/php.12892. Epub 2018 Mar 30. Photochem Photobiol. 2018. PMID: 29441583 Review.
-
Enzymes of Penicillium roqueforti involved in the biosynthesis of cheese flavor.CRC Crit Rev Food Sci Nutr. 1976 Nov;8(2):191-228. doi: 10.1080/10408397609527222. CRC Crit Rev Food Sci Nutr. 1976. PMID: 21770 Review.
References
-
- Sorigue D., Legeret B., Cuine S., Blangy S., Moulin S., Billon E., Richaud P., Brugiere S., Coute Y., Nurizzo D., Muller P., Brettel K., Pignol D., Arnoux P., Li-Beisson Y., Peltier G., Beisson F., Science 2017, 357, 903–907. - PubMed
-
- Schmermund L., Jurkas V., Ozgen F. F., Barone G. D., Büchsenschütz H. C., Winkler C. K., Schmidt S., Kourist R., Kroutil W., ACS Catal. 2019, 9, 4115–4144.
-
- Alphand V., van Berkel W. J. H., Jurkas V., Kara S., Kourist R., Kroutil W., Mascia F., Nowaczyk M. M., Paul C. E., Schmidt S., Spasic J., Tamagnini P., Winkler C. K., ChemPhotoChem 2023, 7, e202200325.
-
- Taylor A., Heyes D. J., Scrutton N. S., Curr. Opin. Struct. Biol. 2022, 77, 102491. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

