This is a preprint.
Role of the SAF-A SAP domain in X inactivation, transcription, splicing, and cell proliferation
- PMID: 39314300
- PMCID: PMC11419091
- DOI: 10.1101/2024.09.09.612041
Role of the SAF-A SAP domain in X inactivation, transcription, splicing, and cell proliferation
Update in
-
Role of the SAF-A/HNRNPU SAP domain in X chromosome inactivation, nuclear dynamics, transcription, splicing, and cell proliferation.PLoS Genet. 2025 Jun 10;21(6):e1011719. doi: 10.1371/journal.pgen.1011719. eCollection 2025 Jun. PLoS Genet. 2025. PMID: 40493679 Free PMC article.
Abstract
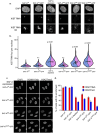
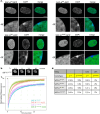
SAF-A is conserved throughout vertebrates and has emerged as an important factor regulating a multitude of nuclear functions, including lncRNA localization, gene expression, and splicing. SAF-A has several functional domains, including an N-terminal SAP domain that binds directly to DNA. Phosphorylation of SAP domain serines S14 and S26 are important for SAF-A localization and function during mitosis, however whether these serines are involved in interphase functions of SAF-A is not known. In this study we tested for the role of the SAP domain, and SAP domain serines S14 and S26 in X chromosome inactivation, protein dynamics, gene expression, splicing, and cell proliferation. Here we show that the SAP domain serines S14 and S26 are required to maintain XIST RNA localization and polycomb-dependent histone modifications on the inactive X chromosome in female cells. In addition, we present evidence that an Xi localization signal resides in the SAP domain. We found that that the SAP domain is not required to maintain gene expression and plays only a minor role in mRNA splicing. In contrast, the SAF-A SAP domain, in particular serines S14 and S26, are required for normal protein dynamics, and to maintain normal cell proliferation. We propose a model whereby dynamic phosphorylation of SAF-A serines S14 and S26 mediates rapid turnover of SAF-A interactions with DNA during interphase.
Figures




References
-
- Skalska L., et al. , Nascent RNA antagonizes the interac1on of a set of regulatory proteins with chroma1n. Mol Cell, 2021. 81(14): p. 2944–2959 e10. - PubMed
-
- Fackelmayer F.O. and Richter A., Purifica1on of two isoforms of hnRNP-U and characteriza1on of their nucleic acid binding ac1vity. Biochemistry, 1994. 33(34): p. 10416–22. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
