This is a preprint.
High-quality peptide evidence for annotating non-canonical open reading frames as human proteins
- PMID: 39314370
- PMCID: PMC11419116
- DOI: 10.1101/2024.09.09.612016
High-quality peptide evidence for annotating non-canonical open reading frames as human proteins
Abstract
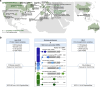
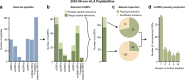
A major scientific drive is to characterize the protein-coding genome as it provides the primary basis for the study of human health. But the fundamental question remains: what has been missed in prior genomic analyses? Over the past decade, the translation of non-canonical open reading frames (ncORFs) has been observed across human cell types and disease states, with major implications for proteomics, genomics, and clinical science. However, the impact of ncORFs has been limited by the absence of a large-scale understanding of their contribution to the human proteome. Here, we report the collaborative efforts of stakeholders in proteomics, immunopeptidomics, Ribo-seq ORF discovery, and gene annotation, to produce a consensus landscape of protein-level evidence for ncORFs. We show that at least 25% of a set of 7,264 ncORFs give rise to translated gene products, yielding over 3,000 peptides in a pan-proteome analysis encompassing 3.8 billion mass spectra from 95,520 experiments. With these data, we developed an annotation framework for ncORFs and created public tools for researchers through GENCODE and PeptideAtlas. This work will provide a platform to advance ncORF-derived proteins in biomedical discovery and, beyond humans, diverse animals and plants where ncORFs are similarly observed.
Keywords: GENCODE; Human Proteome Project; Ribo-seq; immunopeptidomics; mass spectrometry; microproteins; non-canonical ORFs; proteomics; translation.
Conflict of interest statement
Declaration of interests J.R.P. has received research honoraria from Novartis Biosciences and is a paid consultant for ProFound Therapeutics. J.G.A. is a paid consultant for Enara Bio and Moderna. J.L.A. is an advisor to Microneedle Solutions. T.F.M. is a consultant for and holds equity in Velia Therapeutics. J.S.W. is an advisor and holds equity in Velia Therapeutics. G.M. is co-founder and CSO of OHMX.bio. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, Seer and PrognomIQ. N.T.I. hold equity in Velia Therapeutics and holds equity and serves as a scientific advisor to Tevard Biosciences. P.F. is a member of the scientific advisory board of Infinitopes. A.-R. C. is a member of the advisory board of ProFound Therapeutics.
Figures




References
-
- Frankish A. et al. GENCODE: reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res. 51, D942–D949 (2022).
-
- Consortium T. U. et al. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2022).
-
- Ouspenskaia T. et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nature Biotechnology 40, 209–217 (2022).
Publication types
Grants and funding
- R01 GM136849/GM/NIGMS NIH HHS/United States
- U19 AG023122/AG/NIA NIH HHS/United States
- R01 GM087221/GM/NIGMS NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- K08 CA263552/CA/NCI NIH HHS/United States
- P01 CA206978/CA/NCI NIH HHS/United States
- R01 AT012826/AT/NCCIH NIH HHS/United States
- S10 OD026936/OD/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- K01 CA249038/CA/NCI NIH HHS/United States
- U01 CA271402/CA/NCI NIH HHS/United States
- U24 CA270823/CA/NCI NIH HHS/United States
- U24 HG003345/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
