This is a preprint.
Abnormal hyperactivity of specific striatal ensembles encodes distinct dyskinetic behaviors revealed by high-resolution clustering
- PMID: 39314449
- PMCID: PMC11418934
- DOI: 10.1101/2024.09.06.611664
Abnormal hyperactivity of specific striatal ensembles encodes distinct dyskinetic behaviors revealed by high-resolution clustering
Update in
-
Abnormal hyperactivity of specific striatal ensembles encodes distinct dyskinetic behaviors revealed by high-resolution clustering.Cell Rep. 2025 Jul 22;44(7):115988. doi: 10.1016/j.celrep.2025.115988. Epub 2025 Jul 9. Cell Rep. 2025. PMID: 40638389
Abstract
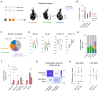
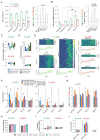
L-DOPA-induced dyskinesia (LID) is a debilitating complication of dopamine replacement therapy in Parkinson's disease and the most common hyperkinetic disorder of basal ganglia origin. Abnormal activity of striatal D1 and D2 spiny projection neurons (SPNs) is critical for LID, yet the link between SPN activity patterns and specific dyskinetic movements remains unknown. To explore this, we developed a novel method for clustering movements based on high-resolution motion sensors and video recordings. In a mouse model of LID, this method identified two main dyskinesia types and pathological rotations, all absent during normal behavior. Using single-cell resolution imaging, we found that specific sets of both D1 and D2-SPNs were abnormally active during these pathological movements. Under baseline conditions, the same SPN sets were active during behaviors sharing physical features with LID movements. These findings indicate that ensembles of behavior-encoding D1- and D2-SPNs form new combinations of hyperactive neurons mediating specific dyskinetic movements.
Keywords: L-DOPA-induced dyskinesia; accelerometer; calcium imaging; freely-moving mouse behavior; inertial measurement units; striatal activity; unsupervised behavioral clustering.
Conflict of interest statement
Competing Interests: The authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
